4YFK
 
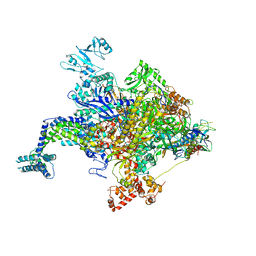 | | Escherichia coli RNA polymerase in complex with squaramide compound 8. | | Descriptor: | 3,5-dimethyl-N-{2-[4-(4-methylbenzyl)piperidin-1-yl]-3,4-dioxocyclobut-1-en-1-yl}-1,2-oxazole-4-sulfonamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.571 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
6F78
 
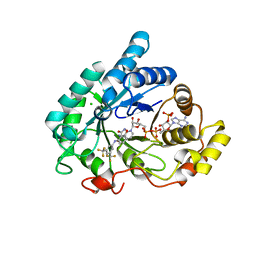 | | Potent and selective Aldo-Keto Reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: Application of a Bioisosteric Scaffold Hopping Approach to Flufenamic acid | | Descriptor: | 4-[[3,5-bis(trifluoromethyl)phenyl]amino]-1,2-benzoxazol-3-one, Aldo-keto reductase family 1 member C3, CHLORIDE ION, ... | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2017-12-07 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Potent and selective aldo-keto reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a bioisosteric scaffold hopping approach to flufenamic acid.
Eur J Med Chem, 150, 2018
|
|
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
9H1E
 
 | |
4YJ5
 
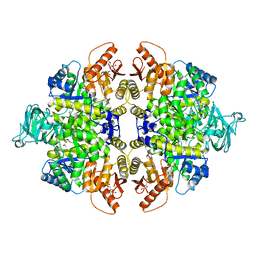 | | Crystal structure of PKM2 mutant | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Liu, J.-S, Wu, C.-W, Wang, W.-C. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Mutations in the PKM2 exon-10 region are associated with reduced allostery and increased nuclear translocation
Commun Biol, 2, 2019
|
|
3IHU
 
 | |
3CWW
 
 | | Crystal Structure of IDE-bradykinin complex | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETATE ION, Insulin-degrading enzyme, ... | | Authors: | Malito, E, Tang, W.J. | | Deposit date: | 2008-04-23 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Molecular Bases for the Recognition of Short Peptide Substrates and Cysteine-Directed Modifications of Human Insulin-Degrading Enzyme
Biochemistry, 47, 2008
|
|
4LJI
 
 | | Crystal structure at 1.5 angstrom resolution of the PsbV2 cytochrome from the cyanobacterium thermosynechococcus elongatus | | Descriptor: | CHLORIDE ION, Cytochrome c-550-like protein, HEME C | | Authors: | Suga, M, Lai, T.-L, Sugiura, M, Shen, J.-R, Boussac, A. | | Deposit date: | 2013-07-04 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Crystal structure at 1.5 angstrom resolution of the PsbV2 cytochrome from the cyanobacterium Thermosynechococcus elongatus
Febs Lett., 587, 2013
|
|
1NES
 
 | | STRUCTURE OF THE PRODUCT COMPLEX OF ACETYL-ALA-PRO-ALA WITH PORCINE PANCREATIC ELASTASE AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | ACETYL-ALA-PRO-ALA, CALCIUM ION, ELASTASE, ... | | Authors: | Meyer Junior, E.F, Radhakrishnan, R, M Cole, G, Presta, L.G. | | Deposit date: | 1995-07-31 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the product complex of acetyl-Ala-Pro-Ala with porcine pancreatic elastase at 1.65 A resolution.
J.Mol.Biol., 189, 1986
|
|
1WWY
 
 | | Solution structure of the DUF1000 domain of a thioredoxin-like protein 1 | | Descriptor: | Thioredoxin-like protein 1 | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal DUF1000 domain of the human thioredoxin-like 1 protein.
Proteins, 78, 2010
|
|
5Y8C
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloranyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
1X3N
 
 | | Crystal structure of AMPPNP bound Propionate kinase (TdcD) from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Propionate kinase | | Authors: | Simanshu, D.K, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of ADP and AMPPNP-bound Propionate Kinase (TdcD) from Salmonella typhimurium: Comparison with Members of Acetate and Sugar Kinase/Heat Shock Cognate 70/Actin Superfamily.
J.Mol.Biol., 352, 2005
|
|
6F2U
 
 | | Potent and selective Aldo-Keto Reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a Bioisosteric Scaffold Hopping Approach to Flufenamic acid | | Descriptor: | 3-[(4-methoxyphenyl)methyl]-5-oxidanyl-~{N}-[3-(trifluoromethyl)phenyl]-1,2,3-triazole-4-carboxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2017-11-27 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Potent and selective aldo-keto reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a bioisosteric scaffold hopping approach to flufenamic acid.
Eur J Med Chem, 150, 2018
|
|
7C6H
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in an open-liganded state bound to laminaribiose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
1NRJ
 
 | |
4FLF
 
 | | Structure of three phase partition treated lipase from Thermomyces lanuginosa at 2.15A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrobenzaldehyde, ... | | Authors: | Kumar, M, Mukherjee, J, Sinha, M, Kaur, P, Gupta, M.N, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-14 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Enhancement of stability of a lipase by subjecting to three phase partitioning (TPP): structures of native and TPP-treated lipase from Thermomyces lanuginosa
Sustain Chem Process, 2015
|
|
2VGP
 
 | | Crystal structure of Aurora B kinase in complex with a aminothiazole inhibitor | | Descriptor: | 4-[(5-bromo-1,3-thiazol-2-yl)amino]-N-methylbenzamide, INNER CENTROMERE PROTEIN A, SERINE/THREONINE-PROTEIN KINASE 12-A | | Authors: | Andersen, C.B, Wan, Y, Chang, J.W, Lee, C, Liu, Y, Sessa, F, Villa, F, Nallan, L, Musacchio, A, Gray, N.S. | | Deposit date: | 2007-11-15 | | Release date: | 2008-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Selective Aminothiazole Aurora Kinase Inhibitors
Acs Chem.Biol., 3, 2008
|
|
6LDR
 
 | |
1O6Z
 
 | | 1.95 A resolution structure of (R207S,R292S) mutant of malate dehydrogenase from the halophilic archaeon Haloarcula marismortui (holo form) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Irimia, A, Ebel, C, Madern, D, Richard, S.B, Cosenza, L.W, Zaccai, G, Vellieux, F.M.D. | | Deposit date: | 2002-10-22 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
4AMF
 
 | | Pseudomonas fluorescens PhoX in complex with the substrate analogue AppCp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
4OEW
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-5-iodo-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
6R50
 
 | |
8V03
 
 | | E. coli 70S ribosome with unmodified P/E-tRNAPro(GGG) bound to slippery P-site CCC-C codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-11-16 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
8UZG
 
 | | E. coli 70S ribosome with unmodified e*/E-tRNAPro(GGG) bound to slippery P-site CCC-C codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-11-15 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
3B8X
 
 | | Crystal structure of GDP-4-keto-6-deoxymannose-3-dehydratase (ColD) H188N mutant with bound GDP-perosamine | | Descriptor: | 1,2-ETHANEDIOL, Pyridoxamine 5-phosphate-dependent dehydrase, SODIUM ION, ... | | Authors: | Cook, P.D, Holden, H.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GDP-4-Keto-6-deoxy-D-mannose 3-Dehydratase, Accommodating a Sugar Substrate in the Active Site.
J.Biol.Chem., 283, 2008
|
|
