1C92
 
 | | Endo-Beta-N-Acetylglucosaminidase H, E132A Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C93
 
 | | Endo-Beta-N-Acetylglucosaminidase H, D130N/E132Q Double Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C94
 
 | | REVERSING THE SEQUENCE OF THE GCN4 LEUCINE ZIPPER DOES NOT AFFECT ITS FOLD. | | Descriptor: | RETRO-GCN4 LEUCINE ZIPPER | | Authors: | Mittl, P.R.E, Deillon, C.A, Sargent, D, Liu, N, Klauser, S, Thomas, R.M, Gutte, B, Gruetter, M.G. | | Deposit date: | 1999-07-30 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1C95
 
 | |
1C96
 
 | | S642A:CITRATE COMPLEX OF ACONITASE | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, MITOCHONDRIAL ACONITASE, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1999-07-31 | | Release date: | 1999-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1C97
 
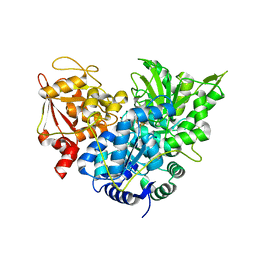 | | S642A:ISOCITRATE COMPLEX OF ACONITASE | | Descriptor: | IRON/SULFUR CLUSTER, ISOCITRIC ACID, MITOCHONDRIAL ACONITASE, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1999-07-31 | | Release date: | 1999-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1C98
 
 | | SOLUTION STRUCTURE OF NEUROMEDIN B | | Descriptor: | NEUROMEDIN B | | Authors: | Lee, S, Kim, Y. | | Deposit date: | 1999-08-01 | | Release date: | 1999-08-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuromedin B by (1)H nuclear magnetic resonance spectroscopy.
FEBS Lett., 460, 1999
|
|
1C99
 
 | |
1C9A
 
 | | SOLUTION STRUCTURE OF NEUROMEDIN B | | Descriptor: | NEUROMEDIN B | | Authors: | Lee, S, Kim, Y. | | Deposit date: | 1999-08-01 | | Release date: | 1999-11-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuromedin B by (1)H nuclear magnetic resonance spectroscopy.
FEBS Lett., 460, 1999
|
|
1C9B
 
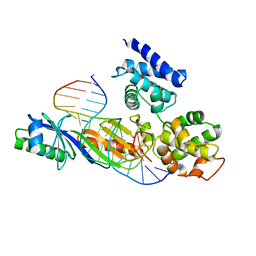 | |
1C9C
 
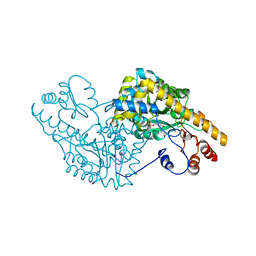 | | ASPARTATE AMINOTRANSFERASE COMPLEXED WITH C3-PYRIDOXAL-5'-PHOSPHATE | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
1C9D
 
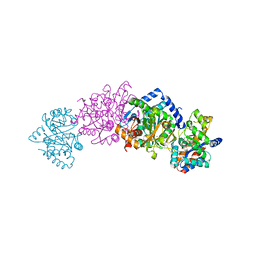 | |
1C9E
 
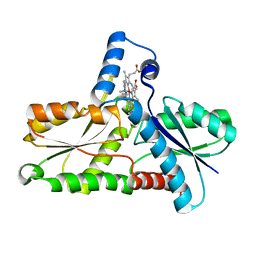 | | STRUCTURE OF FERROCHELATASE WITH COPPER(II) N-METHYLMESOPORPHYRIN COMPLEX BOUND AT THE ACTIVE SITE | | Descriptor: | MAGNESIUM ION, N-METHYLMESOPORPHYRIN CONTAINING COPPER, PROTOHEME FERROLYASE | | Authors: | Lecerof, D, Fodje, M.N, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
1C9F
 
 | |
1C9H
 
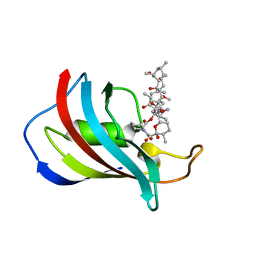 | | CRYSTAL STRUCTURE OF FKBP12.6 IN COMPLEX WITH RAPAMYCIN | | Descriptor: | FKBP12.6, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Deivanayagam, C.C.S, Carson, M, Thotakura, A, Narayana, S.V.L, Chodavarapu, C.S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of FKBP12.6 in complex with rapamycin.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C9I
 
 | |
1C9J
 
 | |
1C9K
 
 | | THE THREE DIMENSIONAL STRUCTURE OF ADENOSYLCOBINAMIDE KINASE/ ADENOSYLCOBINAMIDE PHOSPHATE GUALYLYLTRANSFERASE (COBU) COMPLEXED WITH GMP: EVIDENCE FOR A SUBSTRATE INDUCED TRANSFERASE ACTIVE SITE | | Descriptor: | ADENOSYLCOBINAMIDE KINASE, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thompson, T.B, Thomas, M.G, Esclante-Semerena, J.C, Rayment, I. | | Deposit date: | 1999-08-02 | | Release date: | 1999-08-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase (CobU) complexed with GMP: evidence for a substrate-induced transferase active site.
Biochemistry, 38, 1999
|
|
1C9L
 
 | |
1C9M
 
 | | BACILLUS LENTUS SUBTILSIN (SER 87) N76D/S103A/V104I | | Descriptor: | CALCIUM ION, SERINE PROTEASE, SULFATE ION | | Authors: | Bott, R. | | Deposit date: | 1999-08-02 | | Release date: | 1999-10-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Engineered Bacillus lentus subtilisins having altered flexibility.
J.Mol.Biol., 292, 1999
|
|
1C9N
 
 | |
1C9O
 
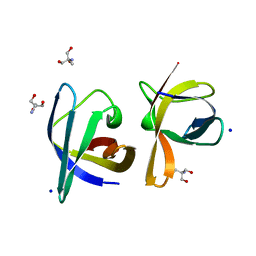 | | CRYSTAL STRUCTURE ANALYSIS OF THE BACILLUS CALDOLYTICUS COLD SHOCK PROTEIN BC-CSP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COLD-SHOCK PROTEIN, SODIUM ION | | Authors: | Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 1999-08-03 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Thermal stability and atomic-resolution crystal structure of the Bacillus caldolyticus cold shock protein.
J.Mol.Biol., 297, 2000
|
|
1C9P
 
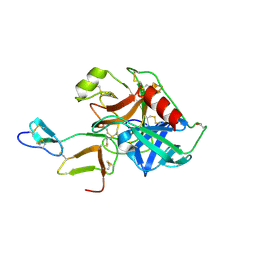 | | COMPLEX OF BDELLASTASIN WITH PORCINE TRYPSIN | | Descriptor: | BDELLASTASIN, CALCIUM ION, TRYPSIN | | Authors: | Rester, U. | | Deposit date: | 1999-08-03 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the complex of the antistasin-type inhibitor bdellastasin with trypsin and modelling of the bdellastasin-microplasmin system.
J.Mol.Biol., 293, 1999
|
|
1C9Q
 
 | |
1C9S
 
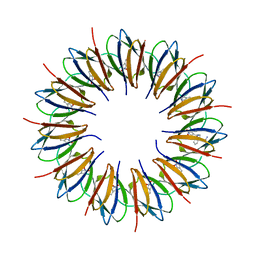 | | CRYSTAL STRUCTURE OF A COMPLEX OF TRP RNA-BINDING ATTENUATION PROTEIN WITH A 53-BASE SINGLE STRANDED RNA CONTAINING ELEVEN GAG TRIPLETS SEPARATED BY AU DINUCLEOTIDES | | Descriptor: | SINGLE STRANDED RNA (55-MER), TRP RNA-BINDING ATTENUATION PROTEIN, TRYPTOPHAN | | Authors: | Antson, A.A, Dodson, E.J, Dodson, G.G, Greaves, R.B, Chen, X.-P, Gollnick, P. | | Deposit date: | 1999-08-03 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA.
Nature, 401, 1999
|
|
