6RZI
 
 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:1(Hydrogen) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-phenylsulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, THIOCYANATE ION | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.095 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
5MS3
 
 | | Kallikrein-related peptidase 8 calcium complex | | Descriptor: | CALCIUM ION, Kallikrein-8 | | Authors: | Debela, M, Magdolen, V, Skala, W, Bode, W, Brandstetter, H, Goettig, P. | | Deposit date: | 2016-12-30 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specificity profiles and antagonistic Ca2+ and Zn2+ regulation of human KLK8/neuropsin activity by modules identified in crystal structures
To Be Published
|
|
6RZM
 
 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:5(Iodine) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(3-iodanylphenyl)sulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Thermodynamic studies of halogen-bond interactions in galectin-3
to be published
|
|
7CU0
 
 | | Crystal structure of Streptomyces albogriseolus flavin-dependent tryptophan 6-halogenase Thal in complex with tryptophan | | Descriptor: | TRYPTOPHAN, Tryptophan 6-halogenase | | Authors: | Chitnumsub, P, Jaruwat, A, Phintha, A, Chaiyen, P. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dissecting the low catalytic capability of flavin-dependent halogenases.
J.Biol.Chem., 296, 2020
|
|
5MVH
 
 | | Glycoside Hydrolase BACCELL_00856 | | Descriptor: | BACCELL_00856 | | Authors: | Munoz-Munoz, J, Cartmell, A, Terrapon, N, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-01-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual active site location and catalytic apparatus in a glycoside hydrolase family.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6RZY
 
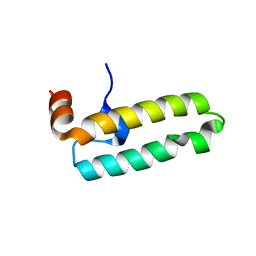 | |
6S00
 
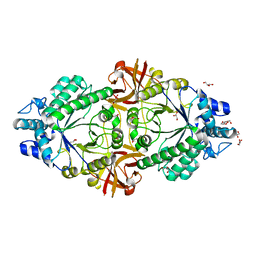 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-acetylneuraminic acid | | Descriptor: | GLYCEROL, N-acetyl-beta-neuraminic acid, TETRAETHYLENE GLYCOL, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
7CDJ
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1A3 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1A3 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.396 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
6S0E
 
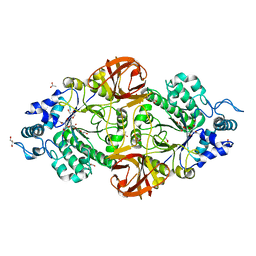 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-Acetyl-2,3-dehydro-2-deoxyneuraminic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ACETATE ION, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-14 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
5MVX
 
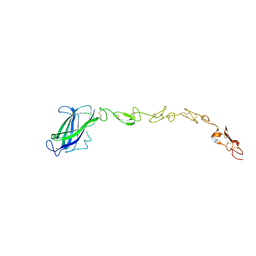 | | Human DLL4 C2-EGF3 | | Descriptor: | Delta-like protein 4, alpha-L-fucopyranose | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
6RFV
 
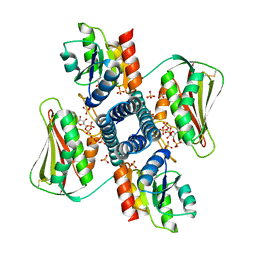 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Response regulator, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
5MW2
 
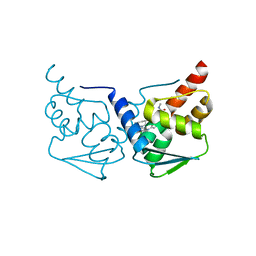 | | CRYSTAL STRUCTURE OF BCL-6 BTB-domain with BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
6RGD
 
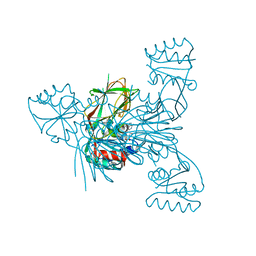 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[4-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]but-3-ynylamino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
5MW7
 
 | | Human Jagged2 C2-EGF3 | | Descriptor: | Protein jagged-2, alpha-L-fucopyranose | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5MT8
 
 | |
5MWB
 
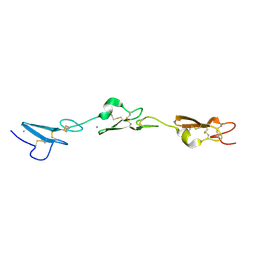 | | Human Notch-2 EGF11-13 | | Descriptor: | CALCIUM ION, Neurogenic locus notch homolog protein 2, alpha-L-fucopyranose, ... | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
6RIM
 
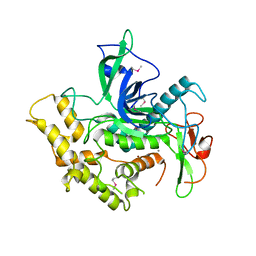 | |
5MWH
 
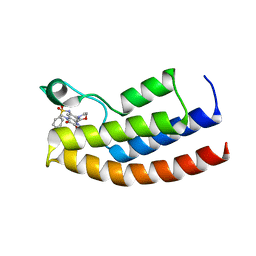 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ089 | | Descriptor: | NITRATE ION, Peregrin, ~{N}-[1,4-dimethyl-7-morpholin-4-yl-2,3-bis(oxidanylidene)quinoxalin-6-yl]-4-(2-methylpropyl)benzenesulfonamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
6RKG
 
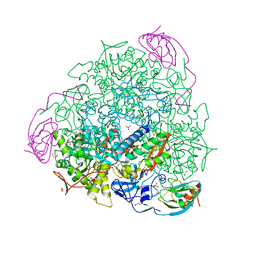 | | 1.32 A RESOLUTION OF SPOROSARCINA PASTEURII UREASE INHIBITED IN THE PRESENCE OF NBPTO AT pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, DIAMIDOPHOSPHATE, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Benini, S, Ciurli, S. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The Impact of pH on Catalytically Critical Protein Conformational Changes: The Case of the Urease, a Nickel Enzyme.
Chemistry, 25, 2019
|
|
5MX9
 
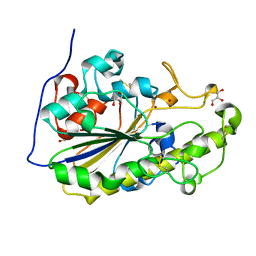 | | High resolution crystal structure of the MCR-2 catalytic domain | | Descriptor: | GLYCEROL, Phosphatidylethanolamine transferase Mcr-2, ZINC ION | | Authors: | Hinchliffe, P, Coates, K, Walsh, T.R, Spencer, J. | | Deposit date: | 2017-01-22 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 1.12 angstrom resolution crystal structure of the catalytic domain of the plasmid-mediated colistin resistance determinant MCR-2.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7C6A
 
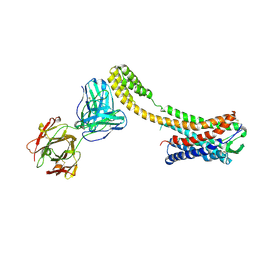 | | Crystal structure of AT2R-BRIL and SRP2070_Fab complex | | Descriptor: | IgG Light Chain, IgG heavy chain, SAR1, ... | | Authors: | Suzuki, M, Miyagi, H, Asada, H, Yasunaga, M, Suno, C, Takahashi, Y, Saito, J, Iwata, S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The discovery of a new antibody for BRIL-fused GPCR structure determination.
Sci Rep, 10, 2020
|
|
5MXJ
 
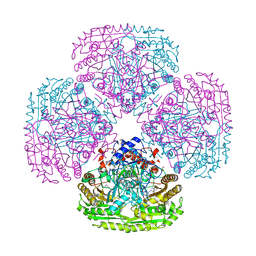 | | Structure of the Y108F mutant of vanillyl alcohol oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Vanillyl-alcohol oxidase | | Authors: | Ewing, T.A, Nguyen, Q.-T, Allan, R.C, Gygli, G, Romero, E, Binda, C, Fraaije, M.W, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2017-01-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two tyrosine residues, Tyr-108 and Tyr-503, are responsible for the deprotonation of phenolic substrates in vanillyl-alcohol oxidase.
J. Biol. Chem., 292, 2017
|
|
6S2X
 
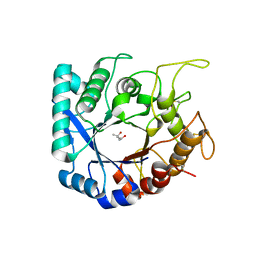 | | Crystal structure of the Legionella pneumophila ChiA C-terminal domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ChiA | | Authors: | Garnett, J.A, Shaw, R. | | Deposit date: | 2019-06-23 | | Release date: | 2020-04-22 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure and functional analysis of the Legionella pneumophila chitinase ChiA reveals a novel mechanism of metal-dependent mucin degradation.
Plos Pathog., 16, 2020
|
|
6RM2
 
 | | Deoxyguanylosuccinate synthase (DgsS) structure with ATP, IMP, Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, INOSINIC ACID, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2019-05-04 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP, IMP, Magnesium at 2.5 Angstrom resolution
To Be Published
|
|
6S3H
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with ADP-AlF4 and (dT)7ds11bp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
