2GLW
 
 | | The solution structure of PHS018 from pyrococcus horikoshii | | Descriptor: | 92aa long hypothetical protein | | Authors: | Coles, M, Hulko, M, Truffault, V, Martin, J, Lupas, A.N. | | Deposit date: | 2006-04-05 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Common evolutionary origin of swapped-hairpin and double-psi beta barrels
Structure, 14, 2006
|
|
2CI7
 
 | | Crystal structure of Dimethylarginine Dimethylaminohydrolase I in complex with Zinc, high pH | | Descriptor: | GLYCINE, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1, ... | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
2GSG
 
 | | Crystal structure of the Fv fragment of a monoclonal antibody specific for poly-glutamine | | Descriptor: | SULFATE ION, monoclonal antibody heavy chain, monoclonal antibody light chain | | Authors: | Li, P, Huey-Tubman, K.E, West Jr, A.P, Bennett, M.J, Bjorkman, P.J. | | Deposit date: | 2006-04-26 | | Release date: | 2007-04-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a polyQ-anti-polyQ complex reveals binding according to a linear lattice model.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2CI6
 
 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase I bound with Zinc low pH | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1, ZINC ION | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inbitors
Structure, 14, 2006
|
|
2GTY
 
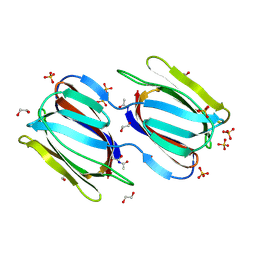 | | Crystal structure of unliganded griffithsin | | Descriptor: | 1,2-ETHANEDIOL, Griffithsin, SULFATE ION | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding.
Structure, 14, 2006
|
|
2GYR
 
 | | Crystal structure of human artemin | | Descriptor: | Neurotrophic factor artemin, isoform 3 | | Authors: | Wang, X.Q. | | Deposit date: | 2006-05-09 | | Release date: | 2006-06-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Artemin Complexed with Its Receptor GFRalpha3: Convergent Recognition of Glial Cell Line-Derived Neurotrophic Factors.
Structure, 14, 2006
|
|
2COQ
 
 | |
2ASY
 
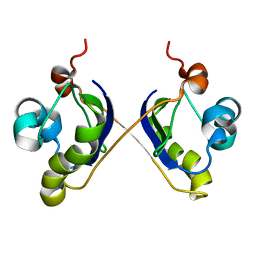 | | Solution Structure of ydhR protein from Escherichia coli | | Descriptor: | Protein ydhR precursor | | Authors: | Revington, M, Semesi, A, Yee, A, Shaw, G.S, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2005-08-24 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Escherichia coli protein ydhR: A putative mono-oxygenase.
Protein Sci., 14, 2005
|
|
2AJU
 
 | |
1PZZ
 
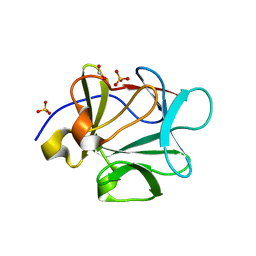 | | Crystal structure of FGF-1, V51N mutant | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-14 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
2GYZ
 
 | | Crystal structure of human artemin | | Descriptor: | neurotrophic factor artemin isoform 3 | | Authors: | Wang, X.Q, Garcia, K.C. | | Deposit date: | 2006-05-10 | | Release date: | 2006-06-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of Artemin Complexed with Its Receptor GFRalpha3: Convergent Recognition of Glial Cell Line-Derived Neurotrophic Factors.
Structure, 14, 2006
|
|
2H2R
 
 | | Crystal structure of the human CD23 Lectin domain, apo form | | Descriptor: | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII)(Immunoglobulin E-binding factor) (CD23 antigen) | | Authors: | Wurzburg, B.A. | | Deposit date: | 2006-05-19 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Changes in the Lectin Domain of CD23, the Low-Affinity IgE Receptor, upon Calcium Binding.
Structure, 14, 2006
|
|
1T4V
 
 | | Crystal Structure Analysis of a novel Oxyguanidine bound to Thrombin | | Descriptor: | Hirudin IIIA, N-ALLYL-5-AMIDINOAMINOOXY-PROPYLOXY-3-CHLORO-N-CYCLOPENTYLBENZAMIDE, Prothrombin | | Authors: | Spurlino, J. | | Deposit date: | 2004-04-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxyguanidines. Part 2: Discovery of a novel orally active thrombin inhibitor through structure-based drug design and parallel synthesis
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
2GUE
 
 | |
1ST4
 
 | | Structure of DcpS bound to m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, YTTRIUM (III) ION, mRNA decapping enzyme | | Authors: | Gu, M, Fabrega, C, Liu, S.W, Liu, H, Kiledjian, M, Lima, C.D. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity
Mol.Cell, 14, 2004
|
|
2H2T
 
 | | CD23 Lectin domain, Calcium 2+-bound | | Descriptor: | CALCIUM ION, Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (Immunoglobulin E-binding factor) (CD23 antigen) | | Authors: | Wurzburg, B.A. | | Deposit date: | 2006-05-19 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Changes in the Lectin Domain of CD23, the Low-Affinity IgE Receptor, upon Calcium Binding.
Structure, 14, 2006
|
|
2EUV
 
 | |
2EWS
 
 | |
2FLN
 
 | | binary complex of catalytic core of human DNA polymerase iota with DNA (template A) | | Descriptor: | DNA polymerase iota, DNA primer strand, DNA template strand | | Authors: | Nair, D.T, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2006-01-06 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site.
Structure, 14, 2006
|
|
2ETW
 
 | |
2EVJ
 
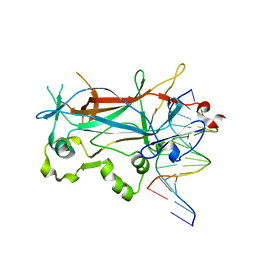 | | Structure of an Ndt80-DNA complex (MSE mutant mA9C) | | Descriptor: | 5'-D(*AP*GP*TP*GP*TP*TP*TP*GP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*CP*AP*AP*AP*CP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Glover, J.N. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Structure, 14, 2006
|
|
2HEV
 
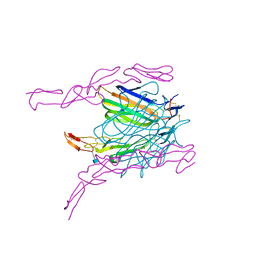 | | Crystal structure of the complex between OX40L and OX40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 4, Tumor necrosis factor receptor superfamily member 4 | | Authors: | Hymowitz, S.G, Compaan, D.M. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The Crystal Structure of the Costimulatory OX40-OX40L Complex.
Structure, 14, 2006
|
|
2HEW
 
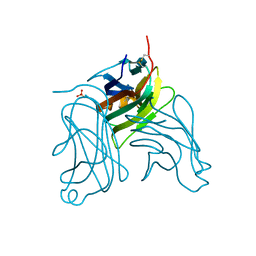 | | The X-ray crystal structure of murine OX40L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Tumor necrosis factor ligand superfamily member 4 | | Authors: | Hymowitz, S.G, Compaan, D.M. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of the Costimulatory OX40-OX40L Complex.
Structure, 14, 2006
|
|
1R78
 
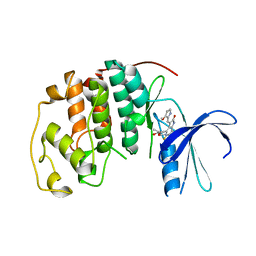 | | CDK2 complex with a 4-alkynyl oxindole inhibitor | | Descriptor: | 4-((3R,4S,5R)-4-AMINO-3,5-DIHYDROXY-HEX-1-YNYL)-5-FLUORO-3-[1-(3-METHOXY-1H-PYRROL-2-YL)-METH-(Z)-YLIDENE]-1,3-DIHYDRO-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Luk, K.-C, Simcox, M.E, Schutt, A, Rowan, K, Thompson, T, Chen, Y, Kammlott, U, DePinto, W, Dunten, P, Dermatakis, A. | | Deposit date: | 2003-10-20 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new series of potent oxindole inhibitors of CDK2
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1R5N
 
 | | Crystal Structure Analysis of sup35 complexed with GDP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
