1W46
 
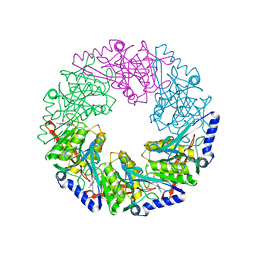 | | P4 protein from Bacteriophage PHI12 in complex with ADP and MG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
3J2V
 
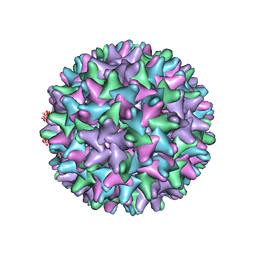 | | CryoEM structure of HBV core | | Descriptor: | PreC/core protein | | Authors: | Yu, X, Jin, L, Jih, J, Shih, C, Zhou, Z.H. | | Deposit date: | 2013-01-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | 3.5 angstrom cryoEM Structure of Hepatitis B Virus Core Assembled from Full-Length Core Protein.
Plos One, 8, 2013
|
|
3JCP
 
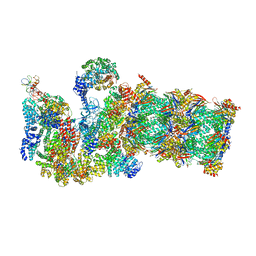 | | Structure of yeast 26S proteasome in M2 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3VK1
 
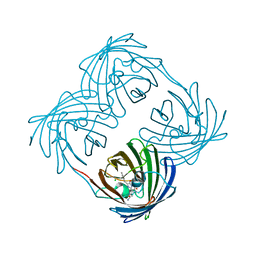 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Wilce, M, Byres, M, Rossjohn, J, Devenish, R.J, Prescott, M. | | Deposit date: | 2011-11-07 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
3J30
 
 | |
3JQX
 
 | | Crystal structure of Clostridium histolyticum colH collagenase collagen binding domain 3 at 2.2 Angstrom resolution in the presence of calcium and cadmium | | Descriptor: | CADMIUM ION, CALCIUM ION, ColH protein | | Authors: | Sakon, J, Philominathan, S.T.L, Bauer, R, Matsushita, O. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
3JRK
 
 | | A putative tagatose 1,6-diphosphate aldolase from Streptococcus pyogenes | | Descriptor: | GLYCEROL, Tagatose 1,6-diphosphate aldolase 2 | | Authors: | Fan, Y, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of a putative tagatose 1,6-diphosphate aldolase from Streptococcus pyogenes
To be Published
|
|
3VM1
 
 | | assimilatory nitrite reductase (Nii3) - N226K mutant - HCO3 complex from tobacco leaf | | Descriptor: | BICARBONATE ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
3J9V
 
 | | Yeast V-ATPase state 3 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3J1U
 
 | | Low affinity dynein microtubule binding domain - tubulin complex | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1, seryl t-RNA synthetase chimera, Tubulin alpha-1B chain, ... | | Authors: | Redwine, W.B, Hernandez-Lopez, R, Zou, S, Huang, J, Reck-Peterson, S.L, Leschziner, A.E. | | Deposit date: | 2012-06-25 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structural basis for microtubule binding and release by dynein.
Science, 337, 2012
|
|
3JT7
 
 | | Structure of neuronal nitric oxide synthase heme domain complexed with N~5~-[2-(propylsulfanyl)ethanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Heme-coordinating inhibitors of neuronal nitric oxide synthase. Iron-thioether coordination is stabilized by hydrophobic contacts without increased inhibitor potency.
J.Am.Chem.Soc., 132, 2010
|
|
3JUL
 
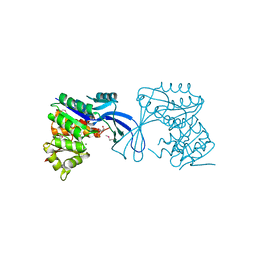 | |
3JUX
 
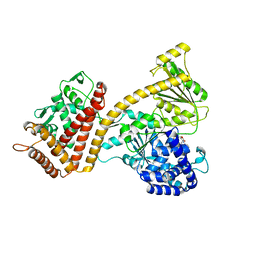 | |
1W48
 
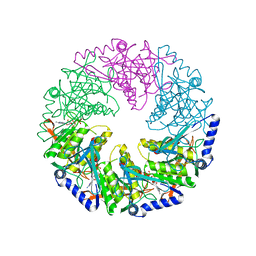 | | P4 protein from Bacteriophage PHI12 in complex with AMPcPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
3J22
 
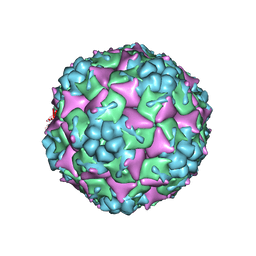 | | The Enterovirus 71 A-particle | | Descriptor: | capsid protein VP0, capsid protein VP1, capsid protein VP3 | | Authors: | Shingler, K.L. | | Deposit date: | 2012-08-13 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | The Enterovirus 71 A-particle Forms a Gateway to Allow Genome Release: A CryoEM Study of Picornavirus Uncoating.
Plos Pathog., 9, 2013
|
|
3J2F
 
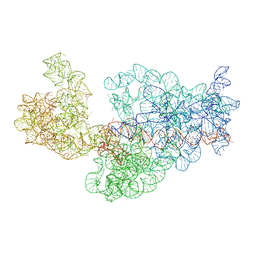 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (17.6 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
3JVY
 
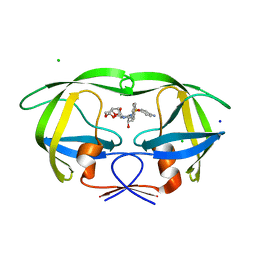 | | HIV-1 Protease Mutant G86A with DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3VRN
 
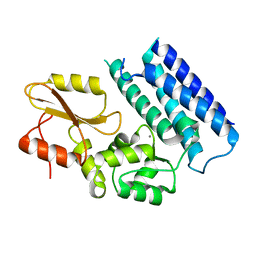 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c | | Descriptor: | CALCIUM ION, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c
J.Biochem., 152, 2012
|
|
3VRU
 
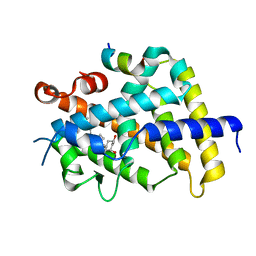 | | VDR ligand binding domain in complex with 2-Methylidene-19,24-dinor-1alpha,25-dihydroxy vitaminD3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R)-5-hydroxy-5-methylhexan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Yoshimoto, N, Inaba, Y, Itoh, T, Nakabayashi, M, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
3JWE
 
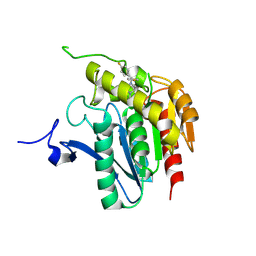 | | Crystal structure of human mono-glyceride lipase in complex with SAR629 | | Descriptor: | 1-[bis(4-fluorophenyl)methyl]-4-(1H-1,2,4-triazol-1-ylcarbonyl)piperazine, MGLL protein | | Authors: | Bertrand, T, Auge, F, Houtmann, J, Rak, A, Vallee, F, Mikol, V, Berne, P.F, Michot, N, Cheuret, D, Hoornaert, C, Mathieu, M. | | Deposit date: | 2009-09-18 | | Release date: | 2010-01-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for human monoglyceride lipase inhibition.
J.Mol.Biol., 396, 2010
|
|
3VPK
 
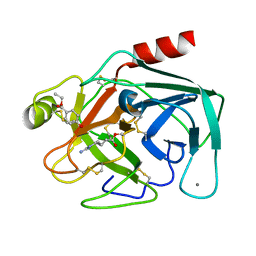 | | Crystal Structure of 6-Guanidinohexanoyl Trypsin | | Descriptor: | 6-carbamimidamidohexanoic acid, CALCIUM ION, Cationic trypsin, ... | | Authors: | Masuda, Y, Nitanai, Y, Mizutani, R, Noguchi, S. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of 6-guanidinohexanoyl trypsin near the optimum pH reveals the acyl-enzyme intermediate to be deacylated
Proteins, 2012
|
|
3JWY
 
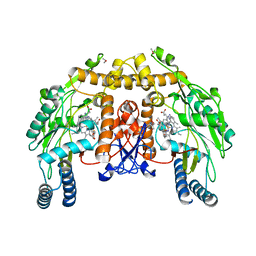 | | Structure of endothelial nitric oxide synthase heme domain complexed with N1-{(3'R,4'S)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLIC ACID, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3J5V
 
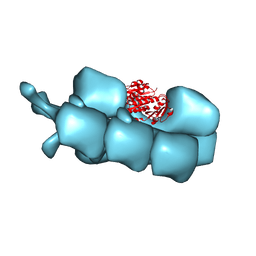 | | PhuZ201 filament | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PhuZ201 subunit | | Authors: | Zehr, E.A. | | Deposit date: | 2013-11-20 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | The Structure and Assembly Mechanism of a Novel Three-Stranded Tubulin Filament that Centers Phage DNA
Structure, 22, 2014
|
|
3J67
 
 | | Structural mechanism of the dynein powerstroke (post-powerstroke state) | | Descriptor: | Dynein motor domain | | Authors: | Lin, J, Okada, K, Raytchev, M, Smith, M.C, Nicastro, D. | | Deposit date: | 2013-12-22 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (34 Å) | | Cite: | Structural mechanism of the dynein power stroke.
Nat.Cell Biol., 16, 2014
|
|
3J6P
 
 | | Pseudo-atomic model of dynein microtubule binding domain-tubulin complex based on a cryoEM map | | Descriptor: | Dynein heavy chain, cytoplasmic, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Uchimura, S, Fujii, T, Takazaki, H, Ayukawa, R, Nishikawa, Y, Minoura, I, Hachikubo, Y, Kurisu, G, Sutoh, K, Kon, T, Namba, K, Muto, E. | | Deposit date: | 2014-03-20 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | A flipped ion pair at the dynein-microtubule interface is critical for dynein motility and ATPase activation
J.Cell Biol., 208, 2015
|
|
