4HTU
 
 | | Structure of 5-chlorouracil modified A:U base pair | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*(UCL)P*CP*GP*CP*G)-3'), GLYCEROL, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
3EIH
 
 | | Crystal structure of S.cerevisiae Vps4 in the presence of ATPgammaS | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gonciarz, M.D, Whitby, F.G, Eckert, D.M, Kieffer, C, Heroux, A, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-09-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Biochemical and structural studies of yeast vps4 oligomerization.
J.Mol.Biol., 384, 2008
|
|
4HMP
 
 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae TIGR4 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, GLYCEROL, ... | | Authors: | Cheng, W, Li, Q, Jiang, Y.-L, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2012-10-18 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Streptococcus pneumoniae PiaA and Its Complex with Ferrichrome Reveal Insights into the Substrate Binding and Release of High Affinity Iron Transporters
Plos One, 8, 2013
|
|
9GRO
 
 | | Crystal structure of the engineered C-terminal phosphatase domain from Saccharomyces cerevisiae Vip1 in complex with 1,5-InsP8 (phosphatase dead mutant, loop deletion residues 848-918) | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], (2S)-hexane-1,2,6-triol, 1,2-ETHANEDIOL, ... | | Authors: | Raia, P, Hothorn, M. | | Deposit date: | 2024-09-11 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A small signaling domain controls PPIP5K phosphatase activity in phosphate homeostasis.
Nat Commun, 16, 2025
|
|
1VF8
 
 | |
5A3H
 
 | | 2-DEOXY-2-FLURO-B-D-CELLOBIOSYL/ENZYME INTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | ENDOGLUCANASE, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
7CCZ
 
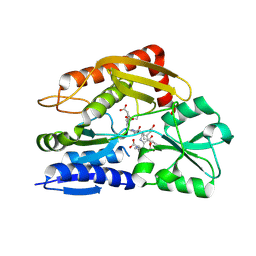 | | Crystal structure of the ES2 intermediate form of human hydroxymethylbilane synthase | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
7UCK
 
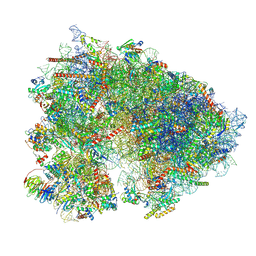 | | 80S translation initiation complex with ac4c(-1) mRNA and Harringtonine | | Descriptor: | 18S rRNA, 28s rRNA, 40S ribosomal protein S10, ... | | Authors: | Yang, R, Arango, D, Sturgill, D, Oberdoerffer, S. | | Deposit date: | 2022-03-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine.
Mol.Cell, 82, 2022
|
|
1J54
 
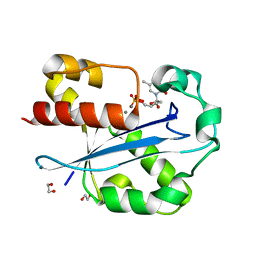 | | Structure of the N-terminal exonuclease domain of the epsilon subunit of E.coli DNA polymerase III at pH 5.8 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
7CD0
 
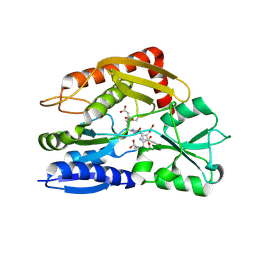 | | Crystal structure of the 2-iodoporphobilinogen-bound ES2 intermediate form of human hydroxymethylbilane synthase | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, 3-[5-(aminomethyl)-4-(carboxymethyl)-2-iodo-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
5T79
 
 | | X-Ray Crystal Structure of a Novel Aldo-keto Reductases for the Biocatalytic Conversion of 3-hydroxybutanal to 1,3-butanediol | | Descriptor: | Aldo-keto Reductase, OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Evdokimova, E, Kudritska, M, Savchenko, A, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl. Environ. Microbiol., 2017
|
|
5YKR
 
 | |
4O14
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
6J9W
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding protein in complex with trehalose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
5TLZ
 
 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with the inhibitor naphthalene 2,6-bisphosphate | | Descriptor: | Fructose-bisphosphate aldolase A, GLYCEROL, naphthalene-2,6-diyl bis[dihydrogen (phosphate)] | | Authors: | Heron, P.W, Sygusch, J. | | Deposit date: | 2016-10-12 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Bisphosphonate Inhibitors of Mammalian Glycolytic Aldolase.
J.Med.Chem., 61, 2018
|
|
1IHH
 
 | | 2.4 ANGSTROM CRYSTAL STRUCTURE OF AN OXALIPLATIN 1,2-D(GPG) INTRASTRAND CROSS-LINK IN A DNA DODECAMER DUPLEX | | Descriptor: | 1R,2R-DIAMINOCYCLOHEXANE, 5'-D(*CP*CP*TP*CP*TP*GP*GP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3', ... | | Authors: | Spingler, B, Whittington, D.A, Lippard, S.J. | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 A crystal structure of an oxaliplatin 1,2-d(GpG) intrastrand cross-link in a DNA dodecamer duplex.
Inorg.Chem., 40, 2001
|
|
6V76
 
 | | Crystal Structure of Human PKM2 in Complex with L-valine | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nandi, S, Dey, M. | | Deposit date: | 2019-12-07 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Biochemical and structural insights into how amino acids regulate pyruvate kinase muscle isoform 2.
J.Biol.Chem., 295, 2020
|
|
6R9W
 
 | | Crystal structure of InhA in complex with AP-124 inhibitor | | Descriptor: | (2~{S})-1-(benzimidazol-1-yl)-3-(2,3-dihydro-1~{H}-inden-5-yloxy)propan-2-ol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Takebayashi, Y, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-04-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of New and Potent InhA Inhibitors as Antituberculosis Agents: Structure-Based Virtual Screening Validated by Biological Assays and X-ray Crystallography.
J.Chem.Inf.Model., 60, 2020
|
|
4ZAY
 
 | | Structure of UbiX E49Q in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
5T5U
 
 | |
9EF2
 
 | | Cryo-EM structure of alpha5beta1 integrin in complex with NeoNectin candidate 2, open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Werther, R, Nguyen, A, Estrada Alamo, K.A, Wang, X, Campbell, M.G. | | Deposit date: | 2024-11-19 | | Release date: | 2025-06-18 | | Last modified: | 2025-09-10 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | De Novo Design of Integrin alpha 5 beta 1 Modulating Proteins to Enhance Biomaterial Properties.
Adv Mater, 37, 2025
|
|
3V7N
 
 | |
2WZX
 
 | |
6FET
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-1439 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{4-[(4aS,8aR)-4-[3,4-bis(difluoromethoxy)phenyl]-1-oxo-1,2,4a,5,8,8a-hexahydrophthalazin-2-yl]piperidin-1-yl}-2-oxoethyl)-4,4-dimethylpiperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-01-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-1439
To be published
|
|
1IXN
 
 | | Enzyme-Substrate Complex of Pyridoxine 5'-Phosphate Synthase | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, Pyridoxine 5'-Phosphate Synthase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
