1XRV
 
 | | Crystal Structure of the novel secretory signalling protein from Porcine (SPP-40) at 2.1A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, signal processing protein | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Singh, N, Sharma, S, Singh, T.P. | | Deposit date: | 2004-10-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the novel secretory signalling protein from Porcine (SPP-40) at 2.1A resolution.
TO BE PUBLISHED
|
|
1XRW
 
 | | Solution Structure of a Platinum-Acridine Modified Octamer | | Descriptor: | 1-[2-(ACRIDIN-9-YLAMINO)ETHYL]-1,3-DIMETHYLTHIOUREA-PLATINUM(II)-ETHANE-1,2-DIAMINE, 5'-D(*CP*CP*TP*CP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*GP*AP*GP*G)-3' | | Authors: | Baruah, H, Wright, M.W, Bierbach, U. | | Deposit date: | 2004-10-16 | | Release date: | 2005-05-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structural Study of a DNA Duplex Containing the Guanine-N7 Adduct Formed by a Cytotoxic Platinum-Acridine Hybrid Agent
Biochemistry, 44, 2005
|
|
1XRX
 
 | | Crystal structure of a DNA-binding protein | | Descriptor: | CALCIUM ION, SeqA protein | | Authors: | Guarne, A, Brendler, T, Zhao, Q, Ghirlando, R, Austin, S, Yang, W. | | Deposit date: | 2004-10-16 | | Release date: | 2005-05-10 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a SeqA-N filament: implications for DNA replication and chromosome organization.
Embo J., 24, 2005
|
|
1XRY
 
 | | Crystal structure of Aeromonas proteolytica aminopeptidase in complex with bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Bacterial leucyl aminopeptidase, ZINC ION | | Authors: | Gilboa, R, Rondeau, J.-M, Blumberg, S, Tarnus, C, Shoham, G. | | Deposit date: | 2004-10-17 | | Release date: | 2005-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interactions of Streptomyces griseus Aminopeptidase and Aeromonas proteolytica Aminopeptidase with Bestatin. Structural analysis of homologous enzymes with different binding modes.
To be Published
|
|
1XRZ
 
 | | NMR Structure of a Zinc Finger with Cyclohexanylalanine Substituted for the Central Aromatic Residue | | Descriptor: | ZINC ION, Zinc finger Y-chromosomal protein | | Authors: | Lachenmann, M.J, Ladbury, J.E, Qian, X, Huang, K, Singh, R, Weiss, M.A. | | Deposit date: | 2004-10-17 | | Release date: | 2004-11-30 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solvation and the hidden thermodynamics of a zinc finger probed
by nonstandard repair of a protein crevice
Protein Sci., 13, 2004
|
|
1XS0
 
 | | Structure of the E. coli Ivy protein | | Descriptor: | Inhibitor of vertebrate lysozyme | | Authors: | Abergel, C, Monchois, V, Byrn, D, Lazzaroni, J.C, Claverie, J.M. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure and evolution of the Ivy protein family, unexpected lysozyme inhibitors in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1XS1
 
 | | dCTP deaminase from Escherichia coli in complex with dUTP | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxycytidine triphosphate deaminase, MAGNESIUM ION | | Authors: | Johansson, E, Fano, M, Bynck, J.H, Neuhard, J, Larsen, S, Sigurskjold, B.W, Christensen, U, Willemoes, M. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes
J.Biol.Chem., 280, 2005
|
|
1XS2
 
 | | Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans | | Descriptor: | MAGNESIUM ION, N-Acylamino Acid Racemase | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
1XS3
 
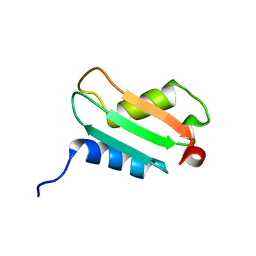 | | Solution Structure Analysis of the XC975 protein | | Descriptor: | hypothetical protein XC975 | | Authors: | Chin, K.-H, Lin, F.-Y, Hu, Y.-C, Sze, K.-H, Lyu, P.-C, Chou, S.-H. | | Deposit date: | 2004-10-18 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Letter to the Editor: NMR structure note - Solution structure of a bacterial BolA-like protein XC975 from a plant pathogen Xanthomonas campestris pv. campestris
J.Biomol.Nmr, 31, 2005
|
|
1XS4
 
 | | dCTP deaminase from Escherichia coli- E138A mutant enzyme in complex with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxycytidine triphosphate deaminase, MAGNESIUM ION | | Authors: | Johansson, E, Fano, M, Bynck, J.H, Neuhard, J, Larsen, S, Sigurskjold, B.W, Christensen, U, Willemoes, M. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes
J.Biol.Chem., 280, 2005
|
|
1XS5
 
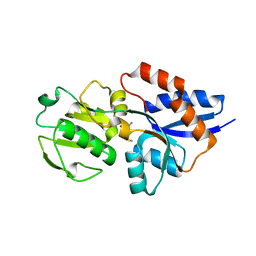 | | The Crystal Structure of Lipoprotein Tp32 from Treponema pallidum | | Descriptor: | METHIONINE, Membrane lipoprotein TpN32 | | Authors: | Deka, R.K, Neil, L, Hagman, K.E, Machius, M, Tomchick, D.R, Brautigam, C.A, Norgard, M.V. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural evidence that the 32-kilodalton lipoprotein (Tp32) of Treponema pallidum is an L-methionine-binding protein
J.Biol.Chem., 279, 2004
|
|
1XS6
 
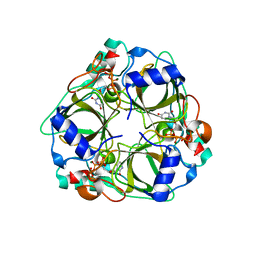 | | dCTP deaminase from Escherichia coli. E138A mutant enzyme in complex with dUTP | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxycytidine triphosphate deaminase, MAGNESIUM ION | | Authors: | Johansson, E, Fano, M, Bynck, J.H, Neuhard, J, Larsen, S, Sigurskjold, B.W, Christensen, U, Willemoes, M. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes
J.Biol.Chem., 280, 2005
|
|
1XS7
 
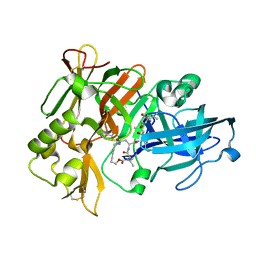 | | Crystal Structure of a cycloamide-urethane-derived novel inhibitor bound to human brain memapsin 2 (beta-secretase). | | Descriptor: | Beta-secretase 1, N-[(4S,5S,7R)-8-({(S)-1-[(BENZYLAMINO)OXOMETHYL]-2-METHYLPROPYL}AMINO)-5-HYDROXY-2,7-DIMETHYL-8-OXO-OCT-4-YL]-(4S,7S)-4 -ISOPROPYL-2,5,9-TRIOXO-1-OXA-3,6,10-TRIAZACYCLOHEXADECANE-7-CARBOXAMIDE | | Authors: | Ghosh, A, Devasamudram, T, Hong, L, DeZutter, C, Xu, X, Weerasena, V, Koelsch, G, Bilcer, G, Tang, J. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of cycloamide-urethane-derived novel inhibitors of human brain memapsin 2 (beta-secretase).
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1XS8
 
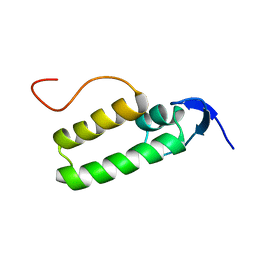 | |
1XS9
 
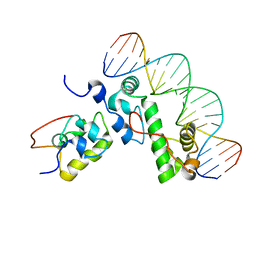 | | A MODEL OF THE TERNARY COMPLEX FORMED BETWEEN MARA, THE ALPHA-CTD OF RNA POLYMERASE AND DNA | | Descriptor: | 5'-D(P*AP*TP*GP*CP*CP*AP*CP*GP*TP*TP*TP*TP*GP*CP*TP*AP*AP*AP*TP*C)-3', 5'-D(P*GP*AP*TP*TP*TP*AP*GP*CP*AP*AP*AP*AP*CP*GP*TP*GP*GP*CP*AP*T)-3', DNA-directed RNA polymerase alpha chain, ... | | Authors: | Dangi, B, Gronenborn, A.M, Rosner, J.L, Martin, R.G. | | Deposit date: | 2004-10-18 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Versatility of the carboxy-terminal domain of the alpha subunit of RNA polymerase in transcriptional activation: use of the DNA contact site as a protein contact site for MarA.
Mol.Microbiol., 54, 2004
|
|
1XSA
 
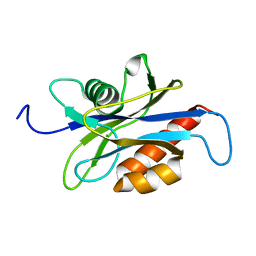 | | Structure of the nudix enzyme AP4A hydrolase from homo sapiens (E63A mutant) | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase | | Authors: | Swarbrick, J.D, Buyya, S, Gunawardana, D, Gayler, K.R, McLennan, A.G, Gooley, P.R. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate-binding Mechanism of Human Ap4A Hydrolase
J.Biol.Chem., 280, 2005
|
|
1XSB
 
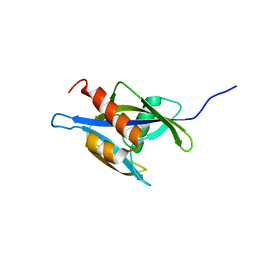 | | Structure of the nudix enzyme AP4A hydrolase from homo sapiens (E63A mutant) in complex with ATP. No ATP restraints included | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase | | Authors: | Swarbrick, J.D, Buyya, S, Gunawardana, D, Gayler, K.R, McLennan, A.G, Gooley, P.R. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate-binding Mechanism of Human Ap4A Hydrolase
J.Biol.Chem., 280, 2005
|
|
1XSC
 
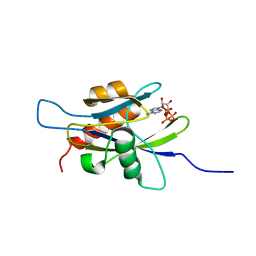 | | Structure of the nudix enzyme AP4A hydrolase from homo sapiens (E63A mutant) in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bis(5'-nucleosyl)-tetraphosphatase | | Authors: | Swarbrick, J.D, Buyya, S, Gunawardana, D, Gayler, K.R, McLennan, A.G, Gooley, P.R. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate-binding Mechanism of Human Ap4A Hydrolase
J.Biol.Chem., 280, 2005
|
|
1XSD
 
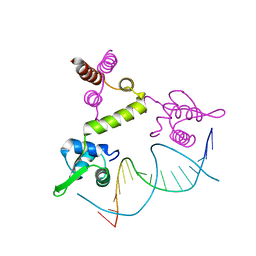 | | Crystal structure of the BlaI repressor in complex with DNA | | Descriptor: | 5'-D(P*TP*AP*CP*TP*AP*CP*AP*TP*AP*TP*GP*TP*AP*GP*TP*A)-3', penicillinase repressor | | Authors: | Safo, M.K, Ko, T.-P, Musayev, F.N, Zhao, Q, Robinson, H, Scarsdale, N, Wang, A.H.-J, Archer, G.L. | | Deposit date: | 2004-10-19 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
1XSE
 
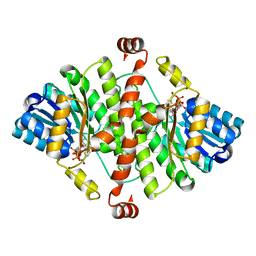 | | Crystal Structure of Guinea Pig 11beta-Hydroxysteroid Dehydrogenase Type 1 | | Descriptor: | 11beta-hydroxysteroid dehydrogenase type 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ogg, D, Elleby, B, Norstrom, C, Stefansson, K, Abrahmsen, L, Oppermann, U, Svensson, S. | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of guinea pig 11beta-hydroxysteroid dehydrogenase type 1 provides a model for enzyme-lipid bilayer interactions
J.Biol.Chem., 280, 2005
|
|
1XSF
 
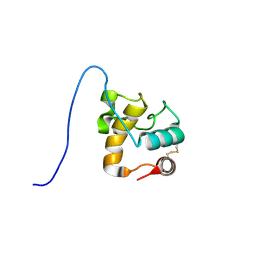 | | Solution structure of a resuscitation promoting factor domain from Mycobacterium tuberculosis | | Descriptor: | Probable resuscitation-promoting factor rpfB | | Authors: | Cohen-Gonsaud, M, Barthe, P, Henderson, B, Ward, J, Roumestand, C, Keep, N.H. | | Deposit date: | 2004-10-19 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structure of a resuscitation-promoting factor domain from Mycobacterium tuberculosis shows homology to lysozymes
Nat.Struct.Mol.Biol., 12, 2005
|
|
1XSG
 
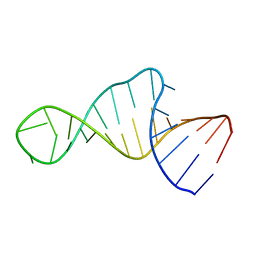 | |
1XSH
 
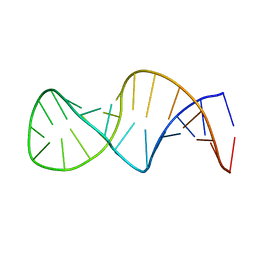 | |
1XSI
 
 | | Structure of a Family 31 alpha glycosidase | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETIC ACID, Putative family 31 glucosidase yicI, ... | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 alpha-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
1XSJ
 
 | | Structure of a Family 31 alpha glycosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative family 31 glucosidase yicI | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 alpha-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
