5Y05
 
 | |
2J6C
 
 | | crystal structure of AFV3-109, a highly conserved protein from crenarchaeal viruses | | Descriptor: | AFV3-109, GLYCEROL | | Authors: | Keller, J, Leulliot, N, Cambillau, C, Campanacci, V, Porciero, S, Prangishvili, D, Cortez, D, Quevillon-Cheruel, S, Van Tilbeurgh, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Afv3-109, a Highly Conserved Protein from Crenarchaeal Viruses.
Virol J., 4, 2007
|
|
6HK1
 
 | | Crystal structure of the Thiazole synthase from Methanothermococcus thermolithotrophicus co-crystallized with Tb-Xo4 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SODIUM ION, ... | | Authors: | Engilberge, S, Wagner, T, Santoni, G, Breyton, C, Shima, S, Franzetti, B, Riobe, F, Maury, O, Girard, E. | | Deposit date: | 2018-09-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Protein crystal structure determination with the crystallophore, a nucleating and phasing agent.
J.Appl.Crystallogr., 52, 2019
|
|
6T5S
 
 | | Apo form of C-type lysozyme from the upper gastrointestinal tract of Opisthocomus hoatzin | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C | | Authors: | Taylor, E.J, Skjot, M, Skov, L.K, Klausen, M, De Maria, L, Gippert, G.P, Turkenburg, J.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-10-17 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-Type Lysozyme from the upper Gastrointestinal Tract of Opisthocomus hoatzin, the Stinkbird.
Int J Mol Sci, 20, 2019
|
|
6T7M
 
 | |
6HN0
 
 | | Complex of Ovine Serum Albumin with diclofenac | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ACETATE ION, ... | | Authors: | Talaj, J.A, Bujacz, A, Bujacz, G. | | Deposit date: | 2018-09-13 | | Release date: | 2019-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins.
Int J Mol Sci, 24, 2023
|
|
7BYU
 
 | | Crystal structure of Acidovorax avenae L-fucose mutarotase (apo form) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, L-fucose mutarotase | | Authors: | Watanabe, Y, Fukui, Y, Watanabe, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Functional and structural characterization of a novel L-fucose mutarotase involved in non-phosphorylative pathway of L-fucose metabolism.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
2JDZ
 
 | | Crystal structure of recombinant Dioclea guianensis lectin complexed with 5-bromo-4-chloro-3-indolyl-a-D-mannose | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CADMIUM ION, CALCIUM ION, ... | | Authors: | Nagano, C.S, Sanz, L, Cavada, B.S, Calvete, J.J. | | Deposit date: | 2007-01-12 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Structural Basis of the Ph-Dependent Dimer-Tetramer Equilibrium Through Crystallographic Analysis of Recombinant Diocleinae Lectins.
Biochem.J., 409, 2008
|
|
2N13
 
 | |
4D4F
 
 | | Mutant P250A of bacterial chalcone isomerase from Eubacterium ramulus | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL | | Authors: | Thomsen, M, Kratzat, H, Hinrichs, W. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus.
Molecules, 27, 2022
|
|
2N0O
 
 | |
5C6J
 
 | | Crystal Structure of Gadolinium derivative of HEWL solved using Free-Electron Laser radiation | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Galli, L, Barends, T.R.M, Son, S.-K, White, T.A, Barty, A, Botha, S, Boutet, S, Caleman, C, Doak, R.B, Nanao, M.H, Nass, K, Shoeman, R.L, Timneanu, N, Santra, R, Schlichting, I, Chapman, H.N. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards phasing using high X-ray intensity.
Iucrj, 2, 2015
|
|
2N9A
 
 | |
5C70
 
 | | The structure of Aspergillus oryzae beta-glucuronidase | | Descriptor: | Glucuronidase | | Authors: | Sun, H.L, Lv, B, Huang, S, Sun, Q.F, Li, C, Jiang, T. | | Deposit date: | 2015-06-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Enhancing the Thermostability of beta-Glucuronidase by Rationally Redesigning the Catalytic Domain Based on Sequence Alignment Strategy
Ind Eng Chem Res, 55, 2016
|
|
2CJ4
 
 | |
2NAA
 
 | | NSD1-PHD_5-C5HCH tandem domain structure | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, ZINC ION | | Authors: | Berardi, A, Quilici, G, Spiliotopoulos, D, Musco, G. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for PHDVC5HCHNSD1-C2HRNizp1 interaction: implications for Sotos syndrome.
Nucleic Acids Res., 44, 2016
|
|
7A70
 
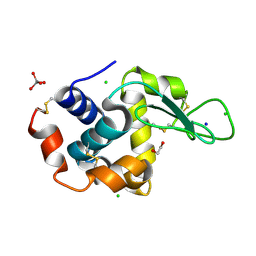 | | HEW lysozyme in complex with Ti(OH)4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Calderone, V, Gigli, L, Ravera, E, Luchinat, C. | | Deposit date: | 2020-08-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the Mechanism of Bioinspired Formation of Inorganic Oxides: Structural Evidence of the Electrostatic Nature of the Interaction between a Mononuclear Inorganic Precursor and Lysozyme.
Biomolecules, 11, 2020
|
|
6SMZ
 
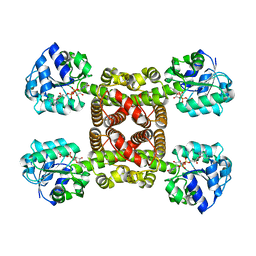 | |
7ADE
 
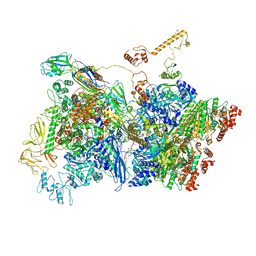 | | Transcription termination complex IVa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6T0F
 
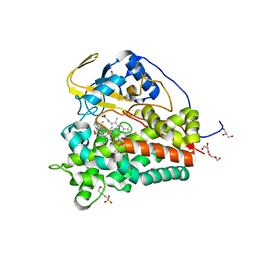 | | Crystal structure of CYP124 in complex with cholest-4-en-3-one | | Descriptor: | (8ALPHA,9BETA)-CHOLEST-4-EN-3-ONE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
3ANW
 
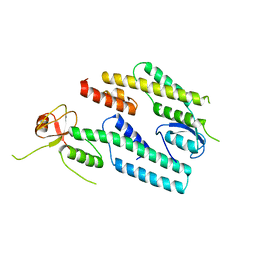 | | A protein complex essential initiation of DNA replication | | Descriptor: | Putative uncharacterized protein | | Authors: | Oyama, T, Shirai, T, Nagasawa, N, Ishino, Y, Morikawa, K. | | Deposit date: | 2010-09-10 | | Release date: | 2011-07-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architectures of archaeal GINS complexes, essential DNA replication initiation factors
Bmc Biol., 9, 2011
|
|
5CA3
 
 | |
2WFB
 
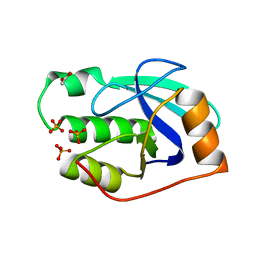 | | High resolution structure of the apo form of the orange protein (ORP) from Desulfovibrio gigas | | Descriptor: | ACETATE ION, PHOSPHATE ION, PUTATIVE UNCHARACTERIZED PROTEIN ORP | | Authors: | Najmudin, S, Bonifacio, C, Duarte, A.G, Pereira, A.S, Moura, I, Moura, J.M, Romao, M.J. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Crystal Structure of the Apo Form of the Orange Protein (Apo-Orp) from Desulfovibrio Gigas
To be Published
|
|
6H8U
 
 | |
6H8W
 
 | |
