1QLY
 
 | | NMR Study of the SH3 Domain From Bruton's Tyrosine Kinase, 20 Structures | | Descriptor: | TYROSINE-PROTEIN KINASE BTK | | Authors: | Tzeng, S.R, Lou, Y.C, Pai, M.T, Chen, C, Chen, S.H, Cheng, J.Y. | | Deposit date: | 1999-09-20 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Human Btk SH3 Domain Complexed with a Proline-Rich Peptide from P120Cbl
J.Biomol.NMR, 16, 2000
|
|
2KWJ
 
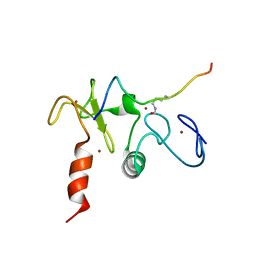 | | Solution structures of the double PHD fingers of human transcriptional protein DPF3 bound to a histone peptide containing acetylation at lysine 14 | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
7Y6C
 
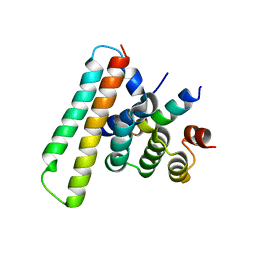 | | Crystal structure of the EscE/EsaG/EsaH complex | | Descriptor: | EscE/YscE/SsaE family type III secretion system needle protein co-chaperone, EscG/YscG/SsaH family type III secretion system needle protein co-chaperone | | Authors: | Zeng, Z.X, Xiao, S.B, Liao, L.J, Zhou, Y.Z. | | Deposit date: | 2022-06-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Secreted in a Type III Secretion System-Dependent Manner, EsaH and EscE Are the Cochaperones of the T3SS Needle Protein EsaG of Edwardsiella piscicida.
Mbio, 13, 2022
|
|
7Y6B
 
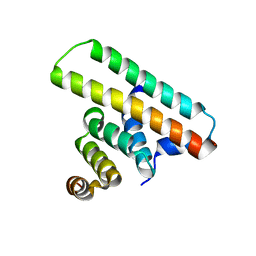 | | Crystal structure of the EscE/EsaH complex | | Descriptor: | EscE/YscE/SsaE family type III secretion system needle protein co-chaperone, EscG/YscG/SsaH family type III secretion system needle protein co-chaperone | | Authors: | Zeng, Z.X, Xiao, S.B, Liao, L.J, Zhou, Y.Z. | | Deposit date: | 2022-06-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Secreted in a Type III Secretion System-Dependent Manner, EsaH and EscE Are the Cochaperones of the T3SS Needle Protein EsaG of Edwardsiella piscicida.
Mbio, 13, 2022
|
|
4RUZ
 
 | |
4RUY
 
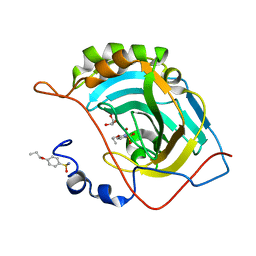 | |
7A20
 
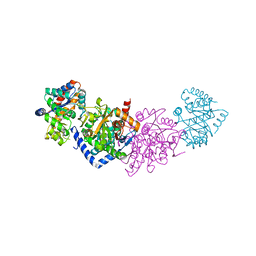 | | Azobenzene-Based Inhibitors for Tryptophan Synthase | | Descriptor: | SODIUM ION, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, Tryptophan synthase alpha chain,Tryptophan synthase beta chain | | Authors: | Rajendran, C, Sterner, R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Towards Photochromic Azobenzene-Based Inhibitors for Tryptophan Synthase.
Chemistry, 27, 2021
|
|
4URI
 
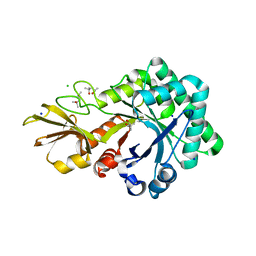 | | Crystal structure of chitinase-like agglutinin RobpsCRA from Robinia pseudoacacia | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHITINASE-RELATED AGGLUTININ, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Henrissat, B, van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2014-06-30 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Carbohydrate Binding Properties of a Plant Chitinase-Like Agglutinin with Conserved Catalytic Machinery.
J.Struct.Biol., 190, 2015
|
|
2QP6
 
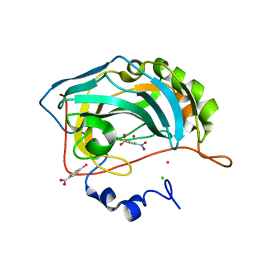 | |
5LXW
 
 | |
7U9I
 
 | | Co-crystal structure of human CARM1 in complex with MT556 inhibitor | | Descriptor: | 7-[5-S-(4-{[(4-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-arginine methyltransferase CARM1, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Perveen, S, Dong, A, Hutchinson, A, Seitova, A, Gibson, E, Hajian, T, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Vedadi, M, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human CARM1 in complex with MT556 inhibitor
To Be Published
|
|
8D41
 
 | | Crystal structure of the human COPB2 WD-domain in complex with OICR-6254 | | Descriptor: | (1R,2R,3S,4S)-3-[4-(4-fluorophenyl)piperazine-1-carbonyl]bicyclo[2.2.1]heptane-2-carboxylic acid, Coatomer subunit beta', GLYCEROL, ... | | Authors: | Zeng, H, Saraon, P, Dong, A, Hutchinson, A, Seitova, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human COPB2 WD-domain in complex with OICR-6254
To Be Published
|
|
4RFP
 
 | |
8D30
 
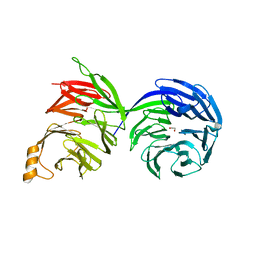 | | Crystal structure of the human COPB2 WD-domains | | Descriptor: | 1,2-ETHANEDIOL, Coatomer subunit beta' | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-31 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human COPB2 WD-domains
To Be Published
|
|
3L82
 
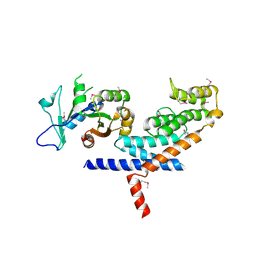 | | X-ray Crystal structure of TRF1 and Fbx4 complex | | Descriptor: | F-box only protein 4, Telomeric repeat-binding factor 1 | | Authors: | Zeng, Z.X, Wang, W, Yang, Y.T, Chen, Y, Yang, X.M, Diehl, J.A, Liu, X.D, Lei, M. | | Deposit date: | 2009-12-29 | | Release date: | 2010-03-09 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Selective Ubiquitination of TRF1 by SCF(Fbx4)
Dev.Cell, 18, 2010
|
|
7NOZ
 
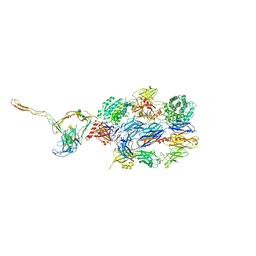 | | Structure of the nanobody stablized properdin bound alternative pathway proconvertase C3b:FB:FP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 alpha chain, Complement C3 beta chain, ... | | Authors: | Lorenzen, J, Pedersen, D.V, Andersen, G.R. | | Deposit date: | 2021-02-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure determination of an unstable macromolecular complex enabled by nanobody-peptide bridging.
Protein Sci., 31, 2022
|
|
1GIK
 
 | | POKEWEED ANTIVIRAL PROTEIN FROM SEEDS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIVIRAL PROTEIN S | | Authors: | Zeng, Z.H, He, X.L, Li, H.M, Hu, Z, Wang, D.C. | | Deposit date: | 2001-02-07 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of pokeweed antiviral protein with well-defined sugars from seeds at 1.8 angstrom resolution
J.Struct.Biol., 141, 2003
|
|
6XHI
 
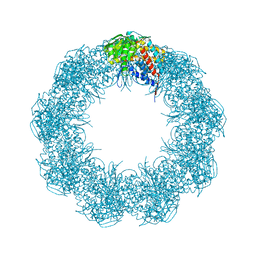 | |
6XHJ
 
 | | Cryo-EM structure of octadecameric TF55 (beta-only) complex from S. solfataricus bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Thermosome subunit beta | | Authors: | Zeng, Y.C, Sobti, M, Stewart, A.G. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural analysis of the Sulfolobus solfataricus TF55beta chaperonin by cryo-electron microscopy
Acta Crystallogr.,Sect.F, 77, 2021
|
|
1JZY
 
 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, ERYTHROMYCIN A, MAGNESIUM ION, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1K01
 
 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, CHLORAMPHENICOL, MAGNESIUM ION, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
8XUD
 
 | |
1JZX
 
 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, CLINDAMYCIN, MAGNESIUM ION, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1JZZ
 
 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, MAGNESIUM ION, ROXITHROMYCIN, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
8XUP
 
 | |
