4FWE
 
 | | Native structure of LSD2 /AOF1/KDM1b in spacegroup of C2221 at 2.13A | | Descriptor: | CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
4G3D
 
 | |
4EFX
 
 | |
4FWF
 
 | | Complex structure of LSD2/AOF1/KDM1b with H3K4 mimic | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.1, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
4G3C
 
 | |
4HLY
 
 | |
4HPH
 
 | | The crystal structure of isomaltulose synthase mutant E295Q from Erwinia rhapontici NX5 in complex with its natural substrate sucrose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HOW
 
 | | The crystal structure of isomaltulose synthase from Erwinia rhapontici NX5 | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-22 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HOZ
 
 | | The crystal structure of isomaltulose synthase mutant D241A from Erwinia rhapontici NX5 in complex with D-glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4I1Q
 
 | | Crystal Structure of hBRAP1 N-BAR domain | | Descriptor: | Bridging integrator 2 | | Authors: | Sanchez-Barrena, M.J. | | Deposit date: | 2012-11-21 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Bin2 Is a Membrane Sculpting N-BAR Protein That Influences Leucocyte Podosomes, Motility and Phagocytosis
Plos One, 7, 2012
|
|
4IGN
 
 | | 2.32 Angstrom X-ray Crystal structure of R47A mutant of human ACMSD | | Descriptor: | 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, ZINC ION | | Authors: | Liu, F, Liu, A. | | Deposit date: | 2012-12-17 | | Release date: | 2014-05-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | Human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD): A structural and mechanistic unveiling.
Proteins, 83, 2015
|
|
4H5E
 
 | | Crystal structure of human FPPS in ternary complex with YS0470 and isopentenyl pyrophosphate | | Descriptor: | Farnesyl pyrophosphate synthase, ISOPENTYL PYROPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Park, J, Lin, Y.-S, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2012-09-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Ternary complex structures of human farnesyl pyrophosphate synthase bound with a novel inhibitor and secondary ligands provide insights into the molecular details of the enzyme's active site closure.
Bmc Struct.Biol., 12, 2012
|
|
4H5C
 
 | | Crystal structure of human FPPS in ternary complex with YS0470 and inorganic phosphate | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Park, J, Lin, Y.-S, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2012-09-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Ternary complex structures of human farnesyl pyrophosphate synthase bound with a novel inhibitor and secondary ligands provide insights into the molecular details of the enzyme's active site closure.
Bmc Struct.Biol., 12, 2012
|
|
4GOB
 
 | | Low pH Crystal Structure of a reconstructed Kaede-type Red Fluorescent Protein, Least Evolved Ancestor (LEA) | | Descriptor: | Kaede-type Fluorescent Protein | | Authors: | Kim, H, Grunkemeyer, T.J, Chen, L, Fromme, R, Wachter, R.M. | | Deposit date: | 2012-08-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Acid-base catalysis and crystal structures of a least evolved ancestral GFP-like protein undergoing green-to-red photoconversion.
Biochemistry, 52, 2013
|
|
4GPZ
 
 | |
4JDD
 
 | |
4JJ3
 
 | |
4H5D
 
 | | Crystal structure of human FPPS in ternary complex with YS0470 and inorganic pyrophosphate | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Park, J, Lin, Y.-S, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2012-09-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Ternary complex structures of human farnesyl pyrophosphate synthase bound with a novel inhibitor and secondary ligands provide insights into the molecular details of the enzyme's active site closure.
Bmc Struct.Biol., 12, 2012
|
|
4J2Q
 
 | |
4JJ0
 
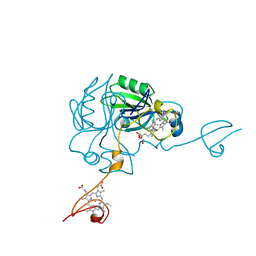 | | Crystal structure of MamP | | Descriptor: | GLYCEROL, HEME C, MamP | | Authors: | Siponen, M, Pignol, D, Arnoux, P. | | Deposit date: | 2013-03-07 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into magnetochrome-mediated magnetite biomineralization.
Nature, 502, 2013
|
|
4JC3
 
 | |
3WDC
 
 | |
3WDD
 
 | |
4J1Q
 
 | |
4J1S
 
 | |
