5SXG
 
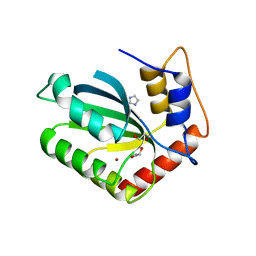 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | 1,3-PROPANDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IMIDAZOLE, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-08-09 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Conformational Switch Regulates the DNA Cytosine Deaminase Activity of Human APOBEC3B.
Sci Rep, 7, 2017
|
|
4ZXX
 
 | |
4FHC
 
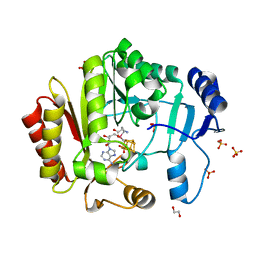 | | Spore photoproduct lyase | | Descriptor: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, SULFATE ION, ... | | Authors: | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
5T2E
 
 | |
5ECU
 
 | |
7G1F
 
 | | Crystal Structure of human FABP4 in complex with 1-(4-methylphenyl)sulfonyl-3-(8-tricyclo[5.2.1.02,6]decanyl)urea, i.e. SMILES [C@@H]12[C@H]3[C@@H]([C@H](C2)C[C@H]1NC(=O)NS(=O)(=O)c1ccc(cc1)C)CCC3 with IC50=1.3 microM | | Descriptor: | 4-methyl-N-{[(3aR,4R,5R,7R,7aR)-octahydro-1H-4,7-methanoinden-5-yl]carbamoyl}benzene-1-sulfonamide, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Stempel, A, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7G1L
 
 | | Crystal Structure of human FABP4 in complex with 6-(1,3-benzodioxol-5-ylmethyl)-3-sulfanyl-1,2,4-triazin-5-ol | | Descriptor: | 6-[(2H-1,3-benzodioxol-5-yl)methyl]-3-sulfanyl-1,2,4-triazin-5-ol, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7DB4
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP012- and glutathione-bound form | | Descriptor: | 1-[(4-fluorophenyl)methyl]-2,2-bis(oxidanylidene)thieno[3,2-c][1,2]thiazin-4-one, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
1QPE
 
 | |
7FVX
 
 | | Crystal Structure of human FABP4 in complex with 4-(1-methyl-2-phenylindol-3-yl)cyclohexane-1-carboxylic acid, i.e. SMILES c1ccc2c(c1)C(=C(N2C)c1ccccc1)[C@@H]1CC[C@@H](CC1)C(=O)O with IC50=0.048 microM | | Descriptor: | (1s,4s)-4-(1-methyl-2-phenyl-1H-indol-3-yl)cyclohexane-1-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Ehler, A, Benz, J, Obst, U, Lerner, C, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3MOH
 
 | |
7FXK
 
 | | Crystal Structure of human FABP4 in complex with 5-benzyl-4-(2,5-dimethylpyrrol-1-yl)-1,2,4-triazole-3-thiol, i.e. SMILES N1(N2C(=CC=C2C)C)C(=NN=C1S)Cc1ccccc1 with IC50=0.462 microM | | Descriptor: | 5-benzyl-4-(2,5-dimethyl-1H-pyrrol-1-yl)-4H-1,2,4-triazole-3-thiol, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kumagai, Y, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5X18
 
 | | Crystal structure of Casein kinase I homolog 1 | | Descriptor: | Casein kinase I homolog 1, GLYCEROL, MALONIC ACID | | Authors: | Kikuchi, M, Shinohara, Y, Ueda, H.R, Umehara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock
Mol. Cell, 67, 2017
|
|
7FYX
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-phenylthiophen-2-yl)carbamoyl]cyclopentene-1-carboxylic acid, i.e. SMILES S1C(=C(C=C1)c1ccccc1)NC(=O)C1=C(CCC1)C(=O)O with IC50=0.201 microM | | Descriptor: | 2-[(3-phenylthiophen-2-yl)carbamoyl]cyclopent-1-ene-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FYJ
 
 | | Crystal Structure of human FABP4 in complex with 5-[(3-chloro-2-methylphenoxy)methyl]-2-phenyl-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one, i.e. SMILES c1(cccc(c1C)Cl)OCC1=CC(=O)N2N=C(c3ccccc3)N=C2N1 with IC50=0.061 microM | | Descriptor: | (8S)-5-[(3-chloro-2-methylphenoxy)methyl]-2-phenyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5EKY
 
 | | Crystal structure of deoxyribose-phosphate aldolase from Escherichia coli (K58E-Y96W mutant) | | Descriptor: | 1,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Deoxyribose-phosphate aldolase | | Authors: | Classen, T, Dick, M, Pietruszka, J, Weiergraeber, O.H. | | Deposit date: | 2015-11-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mechanism-based inhibition of an aldolase at high concentrations of its natural substrate acetaldehyde: structural insights and protective strategies.
Chem Sci, 7, 2016
|
|
4ZXD
 
 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD | | Descriptor: | Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
4DCQ
 
 | | Crystal Structure of the Fab Fragment of 3B5H10, an Antibody-Specific for Extended Polyglutamine Repeats (orthorhombic form) | | Descriptor: | 1,2-ETHANEDIOL, 3B5H10 FAB Heavy Chain, 3B5H10 FAB Light Chain | | Authors: | Peters-Libeu, C.A, Tran, T, Finkbeiner, S, Weisgraber, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
5VH0
 
 | | RHCC in complex with pyrene | | Descriptor: | 1,2-ETHANEDIOL, Tetrabrachion, pyrene | | Authors: | McDougall, M, Francisco, O, Stetefeld, J. | | Deposit date: | 2017-04-12 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Proteinaceous Nano container Encapsulate Polycyclic Aromatic Hydrocarbons.
Sci Rep, 9, 2019
|
|
5IJ6
 
 | | Crystal structure of Enterococcus faecalis lipoate-protein ligase A (lplA-1) in complex with lipoic acid | | Descriptor: | CHLORIDE ION, LIPOIC ACID, Lipoate--protein ligase, ... | | Authors: | Hughes, S.J, Lyle, A.G, Song, J.H, Antoshchenko, T, Park, H. | | Deposit date: | 2016-03-01 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Enterococcus faecalis lipoate-protein ligase A (lplA-1) in complex with lipoic acid
to be published
|
|
1US4
 
 | | PUTATIVE GLUR0 LIGAND BINDING CORE WITH L-GLUTAMATE | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, PUTATIVE GLUR0 LIGAND BINDING CORE | | Authors: | Tahirov, T.H, Inagaki, E, Takahashi, H. | | Deposit date: | 2003-11-18 | | Release date: | 2003-11-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Thermus Thermophilus Putative Periplasmic Glutamate/Glutamine-Binding Protein
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5EJ6
 
 | | EcMenD-ThDP-Mn2+ complex soaked with 2-ketoglutarate for 2min then soaked with isochorismate for 2 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, MANGANESE (II) ION | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | A Thiamine-Dependent Enzyme Utilizes an Active Tetrahedral Intermediate in Vitamin K Biosynthesis
J.Am.Chem.Soc., 138, 2016
|
|
5I3V
 
 | | Crystal structure of BACE1 in complex with aminoquinoline compound 1 | | Descriptor: | (2R)-3-[2-amino-6-(3-methylpyridin-2-yl)quinolin-3-yl]-N-(3,3-dimethylbutyl)-2-methylpropanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2016-02-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fragment-Linking Approach Using (19)F NMR Spectroscopy To Obtain Highly Potent and Selective Inhibitors of beta-Secretase.
J.Med.Chem., 59, 2016
|
|
5ZJF
 
 | | LDHA-MA | | Descriptor: | 5,5'-[(2R,3S)-2,3-dimethylbutane-1,4-diyl]bis(2H-1,3-benzodioxole), L-lactate dehydrogenase A chain | | Authors: | Han, C.W, Jang, S.B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Machilin A Inhibits Tumor Growth and Macrophage M2 Polarization Through the Reduction of Lactic Acid.
Cancers (Basel), 11, 2019
|
|
6JZU
 
 | | The crystal structure of acyl-acyl carrier protein (acyl-ACP) reductase (AAR) in complex with aldehyde deformylating oxygenase (ADO) | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
