1GQ2
 
 | | Malic Enzyme from Pigeon Liver | | Descriptor: | CHLORIDE ION, MALIC ENZYME, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Zhang, H, Liang, T. | | Deposit date: | 2001-11-19 | | Release date: | 2002-05-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of the Pigeon Cytosolic Nadp+ -Dependent Malic Enzyme
Protein Sci., 11, 2002
|
|
1GOK
 
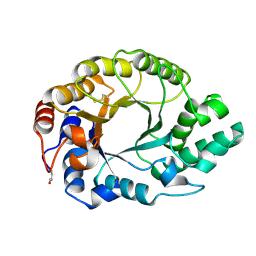 | |
1L0J
 
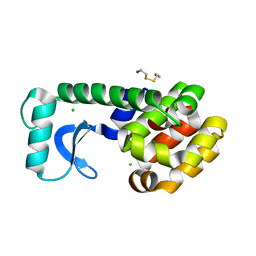 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-02-11 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
3L1S
 
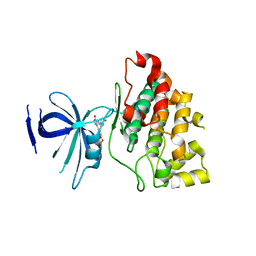 | |
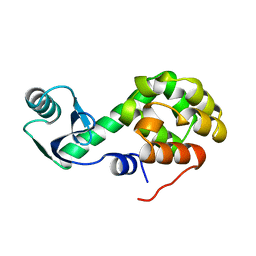
1L51
 
 | |
2ERH
 
 | | Crystal Structure of the E7_G/Im7_G complex; a designed interface between the colicin E7 DNAse and the Im7 immunity protein | | Descriptor: | Colicin E7, Colicin E7 immunity protein | | Authors: | Joachimiak, L.A, Kortemme, T, Stoddard, B.L, Baker, D. | | Deposit date: | 2005-10-24 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational Design of a New Hydrogen Bond Network and at Least a 300-fold Specificity Switch at a Protein-Protein Interface.
J.Mol.Biol., 361, 2006
|
|
5CZY
 
 | | Crystal structure of LegAS4 | | Descriptor: | GLYCEROL, Legionella effector LegAS4, S-ADENOSYLMETHIONINE | | Authors: | Son, J, Hwang, K.Y, Lee, W.C. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Legionella pneumophila type IV secretion system effector LegAS4
Biochem.Biophys.Res.Commun., 465, 2015
|
|
1L18
 
 | |
1L60
 
 | |
4BMJ
 
 | | Structure of the UBZ1and2 tandem of the ubiquitin-binding adaptor protein TAX1BP1 | | Descriptor: | CHLORIDE ION, TAX1-BINDING PROTEIN 1, ZINC ION | | Authors: | Ceregido, M.A, Spinola-Amilibia, M, Buts, L, Rivera, J, Bravo, J, van Nuland, N.A.J. | | Deposit date: | 2013-05-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of Tax1BP1 Ubz1 + 2 Provides Insight Into Target Specificity and Adaptability
J.Mol.Biol., 426, 2014
|
|
1GYE
 
 | | Structure of Cellvibrio cellulosa alpha-L-arabinanase complexed with Arabinohexaose | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A, CHLORIDE ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold.
Nat. Struct. Biol., 9, 2002
|
|
1L67
 
 | |
1L32
 
 | |
1GON
 
 | | b-glucosidase from Streptomyces sp | | Descriptor: | BETA-GLUCOSIDASE, MERCURY (II) ION, SULFATE ION | | Authors: | Guasch, A, Perez-Pons, J.A, Vallmitjana, M, Querol, E, Coll, M. | | Deposit date: | 2001-10-22 | | Release date: | 2002-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Family 1 Beta-Glucosidase from Streptomyces
To be Published
|
|
5D2D
 
 | | Crystal structure of human 14-3-3 zeta in complex with CFTR R-domain peptide pS753-pS768 | | Descriptor: | 14-3-3 protein zeta/delta, CHLORIDE ION, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Stevers, L.M, Leysen, S.F.R, Ottmann, C. | | Deposit date: | 2015-08-05 | | Release date: | 2016-03-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and small-molecule stabilization of the multisite tandem binding between 14-3-3 and the R domain of CFTR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1L74
 
 | |
1GQJ
 
 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with xylobiose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1L94
 
 | |
1GNX
 
 | | b-glucosidase from Streptomyces sp | | Descriptor: | BETA-GLUCOSIDASE, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Guasch, A, Perez-Pons, J.A, Vallmitjana, M, Querol, E, Coll, M. | | Deposit date: | 2001-10-10 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Beta-Glucosidase from Streptomyces
To be Published
|
|
4AFB
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 3 A domain (Epa1to3A) from Candida glabrata in complex with glycerol | | Descriptor: | CALCIUM ION, EPA1P, GLYCEROL | | Authors: | Maestre-Reyna, M, Diderrich, R, Veelders, M.S, Eulenburg, G, Kalugin, V, Brueckner, S, Keller, P, Rupp, S, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Promiscuity and Specificity During Candida Glabrata Invasion of Host Epithelia.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1L56
 
 | |
1GOQ
 
 | |
1GPF
 
 | |
2ER0
 
 | | X-RAY STUDIES OF ASPARTIC PROTEINASE-STATINE INHIBITOR COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, L364,099 | | Authors: | Cooper, J.B, Foundling, S.I, Boger, J, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray studies of aspartic proteinase-statine inhibitor complexes.
Biochemistry, 28, 1989
|
|
5D5H
 
 | | Crystal structure of Mycobacterium tuberculosis Topoisomerase I | | Descriptor: | ACETATE ION, DNA topoisomerase 1, GLYCEROL, ... | | Authors: | Tan, K, Cheng, B, Tse-Dinh, Y.C. | | Deposit date: | 2015-08-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Insights from the Structure of Mycobacterium tuberculosis Topoisomerase I with a Novel Protein Fold.
J.Mol.Biol., 428, 2016
|
|
