7UAZ
 
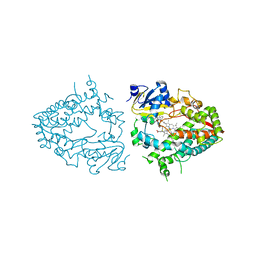 | | Crystal structure of human CYP3A4 with the caged inhibitor | | Descriptor: | (N-[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]-3-{[(pyridin-3-yl)methyl]sulfanyl}propanamide)bis[2-(pyridin-2-yl-kappaN)phenyl-kappaC~1~]iridium, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-03-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Ir(III)-Based Agents for Monitoring the Cytochrome P450 3A4 Active Site Occupancy.
Inorg.Chem., 61, 2022
|
|
9K9N
 
 | | Cryo-EM structure of MjHKU4r-CoV-1 receptor-binding domain complexed with human CD26 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, J.Q. | | Deposit date: | 2024-10-27 | | Release date: | 2025-09-03 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Rational design of human CD26 receptor for a strong neutralizing ability against MjHKU4r-CoV-1 and MERS-CoV
Hlife, 2025
|
|
7UAY
 
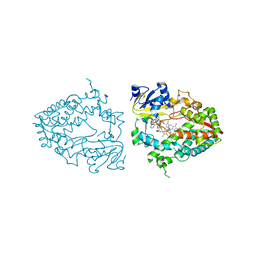 | | Crystal structure of human CYP3A4 with the caged inhibitor | | Descriptor: | (N-[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]-3-{[(pyridin-3-yl)methyl]sulfanyl}propanamide)bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-03-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Ir(III)-Based Agents for Monitoring the Cytochrome P450 3A4 Active Site Occupancy.
Inorg.Chem., 61, 2022
|
|
6QVS
 
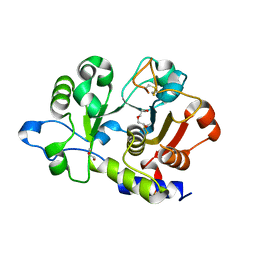 | | Unliganded structure of the human wild type Beta-galactoside alpha-2,6-sialyltransferase 1 (ST6Gal1) | | Descriptor: | Beta-galactoside alpha-2,6-sialyltransferase 1, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Harrus, D, Glumoff, T. | | Deposit date: | 2019-03-04 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unliganded and CMP-Neu5Ac bound structures of human alpha-2,6-sialyltransferase ST6Gal I at high resolution.
J.Struct.Biol., 212, 2020
|
|
4Z2K
 
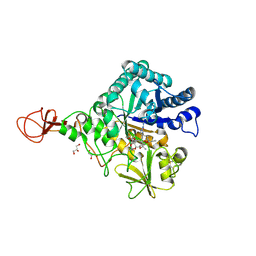 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 32 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {7-[N'-(methylcarbamoyl)carbamimidamido]heptyl}carbamate, Chitinase B, GLYCEROL | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
6UUD
 
 | | Crystal structure of antibody 5D5 in complex with PfCSP N-terminal peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 5D5 Antibody Fab, ... | | Authors: | Thai, E, Scally, S.W, Julien, J.P. | | Deposit date: | 2019-10-30 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A high-affinity antibody against the CSP N-terminal domain lacks Plasmodium falciparum inhibitory activity.
J.Exp.Med., 217, 2020
|
|
7UK1
 
 | |
7AZW
 
 | | Crystal structure of the MIZ1-BTB-domain | | Descriptor: | GLYCEROL, Zinc finger and BTB domain-containing protein 17 isoform X1 | | Authors: | Orth, B, Sander, B, Diederichs, K, Lorenz, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of an atypical interaction site in the BTB domain of the MYC-interacting zinc-finger protein 1.
Structure, 29, 2021
|
|
4Z4W
 
 | | Crystal structure of GII.10 P domain in complex with 4.7mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, beta-L-fucopyranose | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4LKQ
 
 | | Crystal structure of PDE10A2 with fragment ZT017 | | Descriptor: | 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
9H8T
 
 | | Cryo-EM structure of the atovaquone-inhibited Complex III from the Chlorocebus sabaeus respirasome | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-[4-(4-CHLOROPHENYL)CYCLOHEXYLIDENE]-3,4-DIHYDROXY-1(2H)-NAPHTHALENONE, ... | | Authors: | Maclean, A, Muhleip, A. | | Deposit date: | 2024-10-29 | | Release date: | 2025-07-02 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structure, assembly and inhibition of the Toxoplasma gondii respiratory chain supercomplex.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6F91
 
 | | Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
9GMS
 
 | | Mtb PNPase Rv2783c | | Descriptor: | 1-[[4-[4-[[2-phenyl-5-(trifluoromethyl)-1,3-oxazol-4-yl]carbonylamino]phenyl]phenyl]carbonylamino]cyclopentane-1-carboxylic acid, Polyribonucleotide nucleotidyltransferase, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Griesser, T, Sander, P. | | Deposit date: | 2024-08-29 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Selective inhibition of Mycobacterium tuberculosis GpsI unveils a novel strategy to target the RNA metabolism.
Nucleic Acids Res., 53, 2025
|
|
9GMT
 
 | | Mtb PNPase Rv2783c Mutant L328F | | Descriptor: | 1-[[4-[4-[[2-phenyl-5-(trifluoromethyl)-1,3-oxazol-4-yl]carbonylamino]phenyl]phenyl]carbonylamino]cyclopentane-1-carboxylic acid, Polyribonucleotide nucleotidyltransferase, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Griesser, T, Sander, P. | | Deposit date: | 2024-08-29 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Selective inhibition of Mycobacterium tuberculosis GpsI unveils a novel strategy to target the RNA metabolism.
Nucleic Acids Res., 53, 2025
|
|
4GQJ
 
 | | Complex of a binuclear Ruthenium compound D,D-([mu-(11,11')-bi(dppz)-(1,10-phenanthroline)4-Ru2]4+) bound to d(CGTACG) | | Descriptor: | (mu-11,11'-bidipyrido[3,2-a:2',3'-c]phenazine-1kappa~2~N~4~,N~5~:2kappa~2~N~4'~,N~5'~)[tetrakis(1,10-phenanthroline-kappa~2~N~1~,N~10~)]diruthenium, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Boer, D.R, Coll, M. | | Deposit date: | 2012-08-23 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Thread Insertion of a Bis(dipyridophenazine) Diruthenium Complex into the DNA Double Helix by the Extrusion of AT Base Pairs and Cross-Linking of DNA Duplexes.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4YRF
 
 | | Crystal structure of T. cruzi Histidyl-tRNA synthetase in complex with 5-bromopyridin-2(1H)-one (Chem 148) | | Descriptor: | 1,2-ETHANEDIOL, 5-bromopyridin-2(1H)-one, DIMETHYL SULFOXIDE, ... | | Authors: | Koh, C.-Y, Hol, W.G.J. | | Deposit date: | 2015-03-15 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A binding hotspot in Trypanosoma cruzi histidyl-tRNA synthetase revealed by fragment-based crystallographic cocktail screens.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6BQD
 
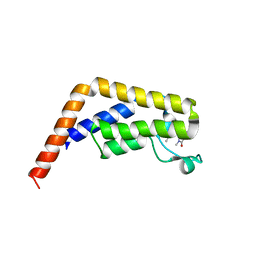 | | TAF1-BD2 bromodomain in complex with (E)-3-(6-(but-2-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide | | Descriptor: | 3-{6-[(2E)-but-2-en-1-yl]-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl}-N,N-dimethylbenzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.136 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
7FZ7
 
 | | Crystal Structure of human FABP4 in complex with 2-[2-[(2-chlorophenyl)methyl]-1-hydroxycyclohexyl]acetic acid, i.e. SMILES [C@]1([C@@H](Cc2c(cccc2)Cl)CCCC1)(CC(=O)O)O with IC50=1.7 microM | | Descriptor: | Fatty acid-binding protein, adipocyte, SULFATE ION, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Dostert, P, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6FJJ
 
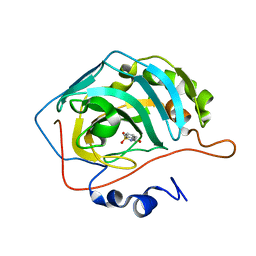 | |
7AT9
 
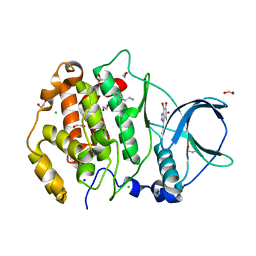 | | Structure of protein kinase ck2 catalytic subunit (csnk2a2 gene product) in complex with the ATP-competitive inhibitor MB002 and the alphaD-pocket ligand 3,4-dichlorophenethylamine | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol, ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
7G01
 
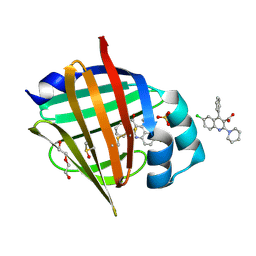 | | Crystal Structure of human FABP5 in complex with 6-chloro-4-phenyl-2-piperidin-1-ylquinoline-3-carboxylic acid, i.e. SMILES n1c(c(c(c2c1ccc(c2)Cl)c1ccccc1)C(=O)O)N1CCCCC1 with IC50=1.1 microM | | Descriptor: | 6-chloranyl-4-phenyl-2-piperidin-1-yl-quinoline-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6V37
 
 | |
7G0F
 
 | | Crystal Structure of human FABP4 in complex with 6-[(3-methoxycarbonyl-4-thiophen-2-ylthiophen-2-yl)carbamoyl]cyclohex-3-ene-1-carboxylic acid, i.e. SMILES C1(=C(NC(=O)[C@H]2[C@@H](C(=O)O)CC=CC2)SC=C1C1=CC=CS1)C(=O)OC with IC50=1.1 microM | | Descriptor: | (1R,2S)-2-{[(2M)-4'-(methoxycarbonyl)-4,5-dihydro-2'H,3H-[2,3'-bi-1lambda~4~-thiophen]-5'-yl]carbamoyl}cyclohexane-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5WB5
 
 | | Leishmania IF4E-1 bound to Leishmania 4E-IP1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative eukaryotic translation initiation factor eIF-4E, Uncharacterized protein | | Authors: | Leger-Abraham, M, Meleppattu, S, Arthanari, H, Zinoviev, A, Boeszoermenyi, A, Wagner, G, Shapira, M. | | Deposit date: | 2017-06-27 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for LeishIF4E-1 modulation by an interacting protein in the human parasite Leishmania major.
Nucleic Acids Res., 46, 2018
|
|
6V3I
 
 | |
