6UME
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-(pyridin-3-yl)-N-(5-{4-[(5-{[(pyridin-3-yl)acetyl]amino}-1,3,4-thiadiazol-2-yl)amino]piperidin-1-yl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
8ZIV
 
 | | wtEP-trypsinogen in Tryp-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Enteropeptidase catalytic light chain, ... | | Authors: | Song, Q.Y, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2024-05-14 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of trypsinogen-engaged EP in Tryp-1 conformation
To Be Published
|
|
6QQX
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-[1-(pyridin-2-ylmethyl)indol-6-yl]-1~{H}-pyrazol-3-amine, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6QR8
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-azanyl-3-[1-[[4-[(4-methylpiperazin-1-yl)methyl]phenyl]methyl]indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
9CRI
 
 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Mu (B.1.621) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
6F08
 
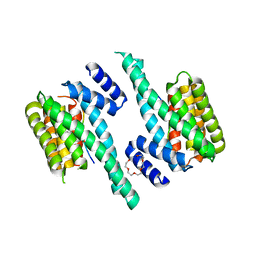 | | 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1) | | Descriptor: | 14-3-3 protein zeta/delta, PENTAETHYLENE GLYCOL, Son of sevenless homolog 1 | | Authors: | Ballone, A, Centorrino, F, Ottmann, C, Guo, S, Leysen, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1).
J. Struct. Biol., 202, 2018
|
|
9CRC
 
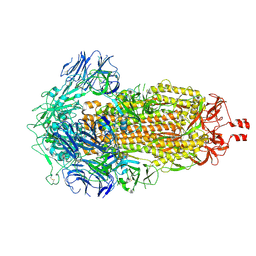 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: B.1 variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
1RKV
 
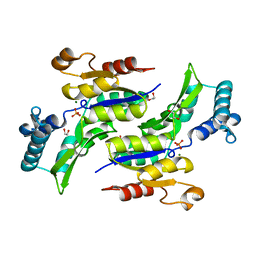 | | Structure of Phosphate complex of ThrH from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Singh, S.K, Yang, K, Subramanian, K, Karthikeyan, S, Huynh, T, Zhang, X, Phillips, M.A, Zhang, H. | | Deposit date: | 2003-11-23 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The thrH Gene Product of Pseudomonas aeruginosa Is a Dual Activity Enzyme with a Novel Phosphoserine:Homoserine Phosphotransferase Activity.
J.Biol.Chem., 279, 2004
|
|
9CRG
 
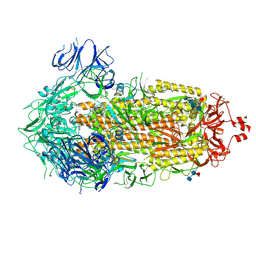 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Gamma (P.1) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Kotecha, A, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
8R4Y
 
 | | Homotrimer for the lumenal domain of Vacuolar Sorting Receptor 1 (VSR1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Vacuolar-sorting receptor 1, alpha-D-mannopyranose, ... | | Authors: | Borges, R.J, Eldahshoury, M.K, Nettleship, J, Bird, L, An, J, Levada, A.J.L, Shah, N, Thomson, M, Fontes, M.R.M, Owens, R, Denecke, J, Postis, V, Uson, I, Goldman, A, De Marcos Lousa, C. | | Deposit date: | 2023-11-14 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The crystal structure of Vacuolar Sorting Receptor 1 (VSR1) lumenal domain reveals a stable domain-swapped trimer and a potential new cargo binding site
To Be Published
|
|
5GMU
 
 | | Crystal structure of chorismate mutase like domain of bifunctional DAHP synthase of Bacillus subtilis in complex with Chlorogenic acid | | Descriptor: | (1R,3R,4S,5R)-3-[3-[3,4-bis(oxidanyl)phenyl]propanoyloxy]-1,4,5-tris(oxidanyl)cyclohexane-1-carboxylic acid, Protein AroA(G), SULFATE ION | | Authors: | Pratap, S, Dev, A, Sharma, V, Yadav, R, Narwal, M, Tomar, S, Kumar, P. | | Deposit date: | 2016-07-16 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Chorismate Mutase-like Domain of DAHPS from Bacillus subtilis Complexed with Novel Inhibitor Reveals Conformational Plasticity of Active Site.
Sci Rep, 7, 2017
|
|
4ZFP
 
 | |
6AX4
 
 | | Plk-1 polo-box domain in complex with histidine N(tau)-cyclized Macrocycle 5b. | | Descriptor: | AMYLAMINE, Serine/threonine-protein kinase PLK1, histidine N(tau)-cyclized Macrocycle 5b | | Authors: | Grant, R.A, Hymel, D, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2017-09-06 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Histidine N( tau )-cyclized macrocycles as a new genre of polo-like kinase 1 polo-box domain-binding inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6AQB
 
 | |
5XZR
 
 | | The atomic structure of SHP2 E76A mutant in complex with allosteric inhibitor 9b | | Descriptor: | 4-(3-phenylphenyl)-N-(2,2,6,6-tetramethylpiperidin-4-yl)-1,3-thiazol-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Li, D, Xie, J, Zhu, J, Liu, C. | | Deposit date: | 2017-07-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric Inhibitors of SHP2 with Therapeutic Potential for Cancer Treatment.
J. Med. Chem., 60, 2017
|
|
7A8Y
 
 | |
3JVR
 
 | | Characterization of the Chk1 allosteric inhibitor binding site | | Descriptor: | (1S)-1-(1H-benzimidazol-2-yl)ethyl (3,4-dichlorophenyl)carbamate, Serine/threonine-protein kinase Chk1 | | Authors: | Chen, P. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Characterization of the CHK1 allosteric inhibitor binding site.
Biochemistry, 48, 2009
|
|
8API
 
 | | THE S VARIANT OF HUMAN ALPHA1-ANTITRYPSIN, STRUCTURE AND IMPLICATIONS FOR FUNCTION AND METABOLISM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-1 ANTITRYPSIN (CHAIN A), ... | | Authors: | Loebermann, H, Tokuoka, R, Deisenhofer, J, Huber, R. | | Deposit date: | 1988-09-08 | | Release date: | 1990-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The S variant of human alpha 1-antitrypsin, structure and implications for function and metabolism.
Protein Eng., 2, 1989
|
|
7ADT
 
 | | Orf virus Apoptosis inhibitor ORFV125 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis inhibitor, Apoptosis regulator BAX, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.210037 Å) | | Cite: | Crystal structures of ORFV125 provide insight into orf virus-mediated inhibition of apoptosis.
Biochem.J., 477, 2020
|
|
5Y4H
 
 | | Human SIRT3 in complex with halistanol sulfate | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, THr-Arg-Ser-GLY-ALY-VAL-MET-ARG-ARG-LEU-ARG-ARG, ... | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Halistanol sulfates I and J, new SIRT1-3 inhibitory steroid sulfates from a marine sponge of the genus Halichondria
J. Antibiot., 71, 2018
|
|
6EV7
 
 | | Structure of E282D A. niger Fdc1 with prFMN in the iminium form | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
8TQ1
 
 | | HIV-1 BG505 Env SOSIP in complex with bovine Fab Bess4 and non-human primate Fab RM20A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bovine Bess4 Fab heavy chain, ... | | Authors: | Ozorowski, G, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
5D2K
 
 | |
6UMC
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-phenyl-N-{5-[(3R)-3-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}oxy)pyrrolidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
7C63
 
 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in an open state (Form I) | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXY BUTANE-1,4-DIOL, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
