9BVR
 
 | | Vitamin K-dependent gamma-carboxylase with factor IX propeptide and partially carboxylated glutamate-rich region and with vitamin K hydroquinone and calcium | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
7E9I
 
 | | Crystal structure of a class I PreQ1 riboswitch aptamer (wild-type) complexed with a cognate ligand-derived photoaffinity probe | | Descriptor: | 2-azanyl-5-[[2-(3-but-3-ynyl-1,2-diazirin-3-yl)ethylamino]methyl]-1,7-dihydropyrrolo[2,3-d]pyrimidin-4-one, 33-mer RNA, SULFATE ION | | Authors: | Numata, T, Schneekloth, J.S. | | Deposit date: | 2021-03-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A chemical probe based on the PreQ 1 metabolite enables transcriptome-wide mapping of binding sites.
Nat Commun, 12, 2021
|
|
5XFC
 
 | | Serial femtosecond X-ray structure of a stem domain of human O-mannose beta-1,2-N-acetylglucosaminyltransferase solved by Se-SAD using XFEL (refined against 13,000 patterns) | | Descriptor: | 4-nitrophenyl beta-D-mannopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Fumiaki, Y, Kato, R, Manya, H. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography
IUCrJ, 4, 2017
|
|
9BVL
 
 | | Vitamin K-dependent gamma-carboxylase with factor X propeptide and glutamate-rich region and with vitamin K hydroquinone | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
9BVQ
 
 | | Vitamin K-dependent gamma-carboxylase with TMG2 propeptide and glutamate-rich region | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
9BVK
 
 | | Vitamin K-dependent gamma-carboxylase with factor IX propeptide and glutamate-rich region and with vitamin K hydroquinone | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
4I3G
 
 | | Crystal Structure of DesR, a beta-glucosidase from Streptomyces venezuelae in complex with D-glucose. | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Beta-glucosidase, ... | | Authors: | Zmudka, M.W, Holden, H.M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of DesR from Streptomyces venezuelae, a beta-glucosidase involved in macrolide activation.
Protein Sci., 22, 2013
|
|
9BVM
 
 | | Vitamin K-dependent gamma-carboxylase with protein C propeptide and glutamate-rich region and with vitamin K hydroquinone | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2024-05-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of vitamin-K-driven gamma-carboxylation at the membrane interface.
Nature, 639, 2025
|
|
6PJ3
 
 | | Crystal structure of the Klebsiella pneumoniae LpxH/JH-LPH-33 complex | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-({4-[3-chloro-5-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5F0A
 
 | | CRYSTAL STRUCTURE OF PVX_084705 WITH BOUND 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine INHIBITOR | | Descriptor: | 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, cGMP-dependent protein kinase, putative | | Authors: | Walker, J.R, Wernimont, A.K, He, H, Seitova, A, Loppnau, P, Sibley, L.D, Graslund, S, Hutchinson, A, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Hui, R, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF PVX_084705 WITH BOUND INHIBITOR
To be published
|
|
5SPN
 
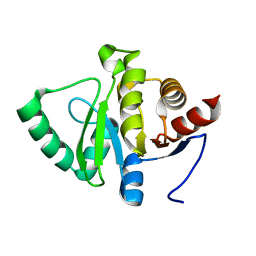 | |
6DXT
 
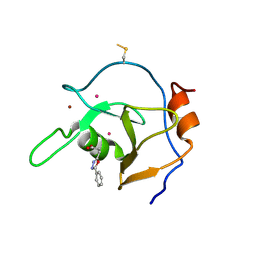 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 3-(5-phenyl-1,3,4-oxadiazol-2-yl)propanoate | | Descriptor: | 1,2-ETHANEDIOL, 3-(5-phenyl-1,3,4-oxadiazol-2-yl)propanoic acid, UNKNOWN ATOM OR ION, ... | | Authors: | Mann, M.K, Harding, R.J, Ravichandran, M, Ferreira de Freitas, R, Franzoni, I, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
7U08
 
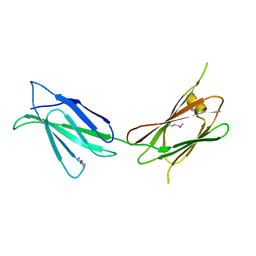 | |
4LXQ
 
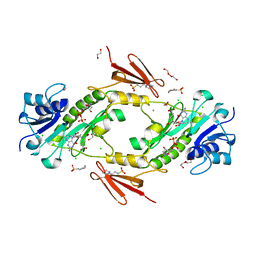 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTdp and 5-N-Formyl-THF | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
7AIY
 
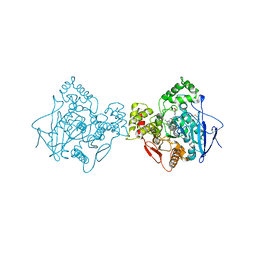 | | Crystal structure of human butyrylcholinesterase in complex with 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide | | Descriptor: | 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide, Cholinesterase | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.937 Å) | | Cite: | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7QTT
 
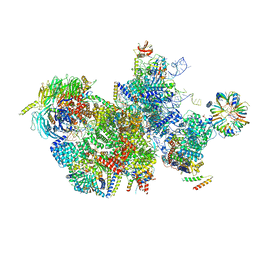 | |
1IXN
 
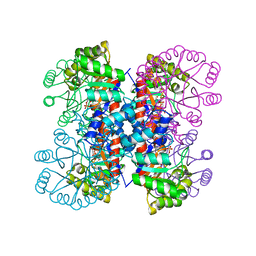 | | Enzyme-Substrate Complex of Pyridoxine 5'-Phosphate Synthase | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, Pyridoxine 5'-Phosphate Synthase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
3W2S
 
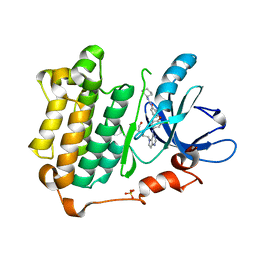 | | EGFR kinase domain with compound4 | | Descriptor: | 1-{3-[2-chloro-4-({5-[2-(2-hydroxyethoxy)ethyl]-5H-pyrrolo[3,2-d]pyrimidin-4-yl}amino)phenoxy]phenyl}-3-cyclohexylurea, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
3KVY
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 1,4-anhydro-D-erythro-pent-1-enitol, SULFATE ION, URACIL, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
8H5S
 
 | | Crystal structure of Rv3400 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Beta-phosphoglucomutase, CHLORIDE ION, ... | | Authors: | Singh, L, Karthikeyan, S, Thakur, K.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization reveals Rv3400 codes for beta-phosphoglucomutase in Mycobacterium tuberculosis.
Protein Sci., 33, 2024
|
|
5XDJ
 
 | | Esculentin-1a(1-21)NH2 | | Descriptor: | Esculentin-1A | | Authors: | Ghosh, A, Bhunia, A. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Membrane perturbing activities and structural properties of the frog-skin derived peptide Esculentin-1a(1-21)NH2 and its Diastereomer Esc(1-21)-1c: Correlation with their antipseudomonal and cytotoxic activity
Biochim. Biophys. Acta, 1859, 2017
|
|
5BT9
 
 | |
6A80
 
 | | Crystal Structure of putative amino acid binding periplasmic ABC transporter protein from Candidatus Liberibacter asiaticus in complex with cystine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Kumar, P, Kesari, P, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-07-05 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structures of a putative periplasmic cystine-binding protein from Candidatus Liberibacter asiaticus: insights into an adapted mechanism of ligand binding.
Febs J., 286, 2019
|
|
1J54
 
 | | Structure of the N-terminal exonuclease domain of the epsilon subunit of E.coli DNA polymerase III at pH 5.8 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
6THY
 
 | | Botulinum neurotoxin A3 Hc domain in complex with GD1a | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BoNT/A3, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2019-11-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of botulinum neurotoxin subtype A3 cell binding domain in complex with GD1a co-receptor ganglioside.
Febs Open Bio, 10, 2020
|
|
