4K6T
 
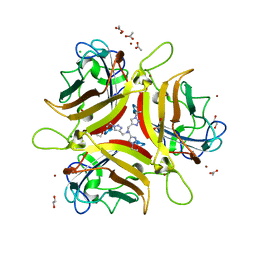 | | Crystal structure of Ad37 fiber knob in complex with trivalent sialic acid inhibitor ME0385 | | Descriptor: | 1,2-ETHANEDIOL, 2,2',2''-[nitrilotris(methanediyl-1H-1,2,3-triazole-4,1-diyl)]triethanol, ACETATE ION, ... | | Authors: | Stehle, T, Bauer, J. | | Deposit date: | 2013-04-16 | | Release date: | 2014-10-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells.
Org.Biomol.Chem., 13, 2015
|
|
3OEY
 
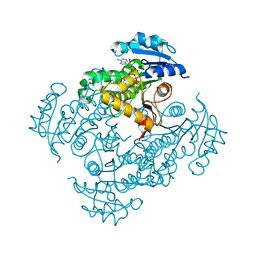 | | Crystal structure of InhA_T266E:NADH complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Molle, V, Gulten, G, Vilcheze, C, Veyron-Churlet, R, Zanella-Cleon, I, Sacchettini, J.C, Jacobs Jr, W.R, Kremer, L. | | Deposit date: | 2010-08-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis.
Mol.Microbiol., 78, 2010
|
|
1UO6
 
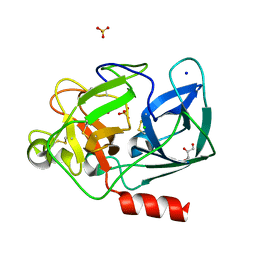 | | PORCINE PANCREATIC ELASTASE/Xe-COMPLEX | | Descriptor: | CHLORIDE ION, ELASTASE 1, GLYCEROL, ... | | Authors: | Mueller-Dieckmann, C, Polentarutti, M, Djinovic-Carugo, K, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2003-09-16 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | On the Routine Use of Soft X-Rays in Macromolecular Crystallography. Part II. Data-Collection Wavelength and Scaling Models
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3OEW
 
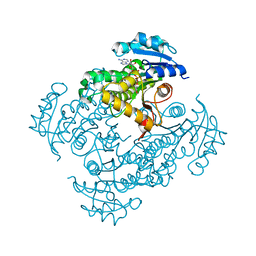 | | Crystal structure of wild-type InhA:NADH complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Molle, V, Gulten, G, Vilcheze, C, Veyron-Churlet, R, Zanella-Cleon, I, Sacchettini, J.C, Jacobs Jr, W.R, Kremer, L. | | Deposit date: | 2010-08-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis.
Mol.Microbiol., 78, 2010
|
|
3OTS
 
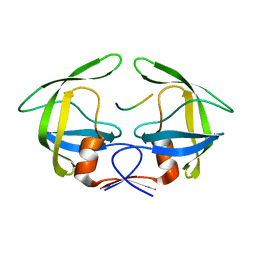 | | MDR769 HIV-1 protease complexed with MA/CA hepta-peptide | | Descriptor: | MA/CA substrate peptide, Multi-drug resistant HIV-1 protease | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-13 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OTY
 
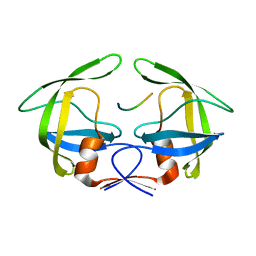 | | MDR769 HIV-1 protease complexed with RT/RH hepta-peptide | | Descriptor: | MDR HIV-1 protease, RT/RH substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OUA
 
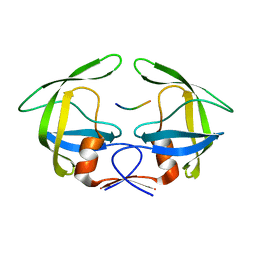 | | MDR769 HIV-1 protease complexed with p1/p6 hepta-peptide | | Descriptor: | HIV-1 protease, p1/p6 substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3P2B
 
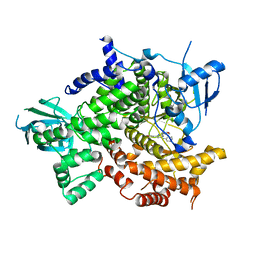 | | Crystal Structure of PI3K gamma with 3-(2-morpholino-6-(pyridin-3-ylamino)pyrimidin-4-yl)phenol | | Descriptor: | 3-[2-morpholin-4-yl-6-(pyridin-3-ylamino)pyrimidin-4-yl]phenol, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Knapp, M.S, Elling, R.A, Ornelas, E. | | Deposit date: | 2010-10-01 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification and structure-activity relationship of 2-morpholino 6-(3-hydroxyphenyl) pyrimidines, a class of potent and selective PI3 kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4K6W
 
 | | Crystal structure of Ad37 fiber knob in complex with trivalent sialic acid inhibitor ME0408 | | Descriptor: | 1,2-ETHANEDIOL, 2,2',2''-[nitrilotris(methanediyl-1H-1,2,3-triazole-4,1-diyl)]triethanol, 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, ... | | Authors: | Stehle, T, Bauer, J. | | Deposit date: | 2013-04-16 | | Release date: | 2014-10-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells.
Org.Biomol.Chem., 13, 2015
|
|
2F7A
 
 | | BenM effector binding domain with its effector, cis,cis-muconate | | Descriptor: | (2Z,4Z)-HEXA-2,4-DIENEDIOIC ACID, ACETATE ION, BENZOIC ACID, ... | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-30 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
1CE1
 
 | | 1.9A STRUCTURE OF THE THERAPEUTIC ANTIBODY CAMPATH-1H FAB IN COMPLEX WITH A SYNTHETIC PEPTIDE ANTIGEN | | Descriptor: | PROTEIN (CAMPATH-1H:HEAVY CHAIN), PROTEIN (CAMPATH-1H:LIGHT CHAIN), PROTEIN (PEPTIDE ANTIGEN) | | Authors: | James, L.C, Hale, G, Waldmann, H, Bloomer, A.C. | | Deposit date: | 1999-03-12 | | Release date: | 1999-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 A structure of the therapeutic antibody CAMPATH-1H fab in complex with a synthetic peptide antigen.
J.Mol.Biol., 289, 1999
|
|
1SUF
 
 | | Carbon Monoxide Dehydrogenase from Carboxydothermus hydrogenoformans-Inactive state | | Descriptor: | Carbon Monoxide Dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
4KZ0
 
 | | Structure of PI3K gamma with Imidazopyridine inhibitors | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, methyl 2-(acetylamino)-1,3-benzothiazole-6-carboxylate | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure guided optimization of a fragment hit to imidazopyridine inhibitors of PI3K.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KZC
 
 | | Structure of PI3K gamma with Imidazopyridine inhibitors | | Descriptor: | N-{6-[6-amino-5-(trifluoromethyl)pyridin-3-yl]imidazo[1,2-a]pyridin-2-yl}acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Knapp, M.S, Elling, E.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure guided optimization of a fragment hit to imidazopyridine inhibitors of PI3K.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2F6G
 
 | | BenM effector binding domain | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator benM, SULFATE ION | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-29 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
1SU6
 
 | | Carbon monoxide dehydrogenase from Carboxydothermus hydrogenoformans: CO reduced state | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
2F6P
 
 | | BenM effector binding domain- SeMet derivative | | Descriptor: | ACETATE ION, HTH-type transcriptional regulator benM, SODIUM ION, ... | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-29 | | Release date: | 2006-10-31 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
6LHO
 
 | | The cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHK
 
 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LJB
 
 | |
4E1A
 
 | | Phosphopantetheine adenylyltransferase from Mycobacterium tuberculosis at 1.62A resolution | | Descriptor: | GLYCEROL, Phosphopantetheine adenylyltransferase | | Authors: | Timofeev, V.I, Smirnova, E.A, Chupova, L.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2012-03-06 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray study of the conformational changes in the molecule of phosphopantetheine adenylyltransferase from Mycobacterium tuberculosis during the catalyzed reaction.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6ZWW
 
 | | Crystal structure of E. coli RNA helicase HrpA in complex with RNA | | Descriptor: | ATP-dependent RNA helicase HrpA, CALCIUM ION, ssRNA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZWX
 
 | | Crystal structure of E. coli RNA helicase HrpA | | Descriptor: | ATP-dependent RNA helicase HrpA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-30 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6LHA
 
 | | The cryo-EM structure of coxsackievirus A16 mature virion | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHQ
 
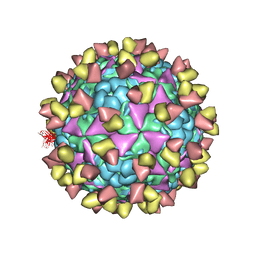 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab NA9D7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
