1QU1
 
 | | CRYSTAL STRUCTURE OF EHA2 (23-185) | | Descriptor: | PROTEIN (INFLUENZA RECOMBINANT HA2 CHAIN) | | Authors: | Chen, J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1999-07-05 | | Release date: | 2000-01-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1YY0
 
 | | Crystal structure of an RNA duplex containing a 2'-amine substitution and a 2'-amide product produced by in-crystal acylation at a C-A mismatch | | Descriptor: | 5'-R(*GP*CP*AP*GP*AP*(A5M)P*UP*UP*AP*AP*AP*UP*CP*UP*GP*C)-3', 5'-R(*GP*CP*AP*GP*AP*(M5M)P*UP*UP*AP*AP*AP*UP*CP*UP*GP*C)-3', CALCIUM ION | | Authors: | Gherghe, C.M, Krahn, J.M, Weeks, K.M. | | Deposit date: | 2005-02-23 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures, reactivity and inferred acylation transition States for 2'-amine substituted RNA.
J.Am.Chem.Soc., 127, 2005
|
|
4A71
 
 | | cytochrome c peroxidase in complex with phenol | | Descriptor: | CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, PHENOL, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Guaiacol and Phenol Bound to a Heme Peroxidase.
FEBS J., 279, 2012
|
|
4A6Z
 
 | | Cytochrome c peroxidase with bound guaiacol | | Descriptor: | CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, Guaiacol, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Nnamchi, C, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Guaiacol and Phenol Bound to a Heme Peroxidase.
FEBS J., 279, 2012
|
|
4A7M
 
 | | cytochrome c peroxidase S81W mutant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of Guaiacol and Phenol Bound to a Heme Peroxidase.
FEBS J., 279, 2012
|
|
1S6V
 
 | | Structure of a cytochrome c peroxidase-cytochrome c site specific cross-link | | Descriptor: | Cytochrome c peroxidase, mitochondrial, Cytochrome c, ... | | Authors: | Guo, M, Bhaskar, B, Li, H, Barrows, T.P, Poulos, T.L. | | Deposit date: | 2004-01-27 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure and characterization of a cytochrome c peroxidase-cytochrome c site-specific cross-link
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1EBE
 
 | | Laue diffraction study on the structure of cytochrome c peroxidase compound I | | Descriptor: | CYTOCHROME C PEROXIDASE, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fulop, V, Phizackerley, R.P, Soltis, S.M, Clifton, I.J, Wakatsuki, S, Erman, J.E, Hajdu, J, Edwards, S.L. | | Deposit date: | 2001-07-25 | | Release date: | 2001-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Laue Diffraction Study on the Structure of Cytochrome C Peroxidase Compound I
Structure, 2, 1994
|
|
4A78
 
 | | cytochrome c peroxidase M119W in complex with guiacol | | Descriptor: | CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, Guaiacol, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2011-11-11 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Guaiacol and Phenol Bound to a Heme Peroxidase.
FEBS J., 279, 2012
|
|
1MBH
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | C-MYB | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
4U5J
 
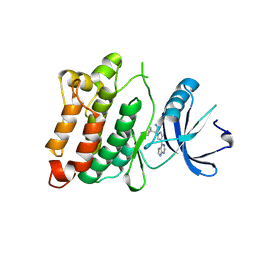 | | C-Src in complex with Ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Chen, Y, Duan, Y, Chen, L. | | Deposit date: | 2014-07-25 | | Release date: | 2014-09-17 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | c-Src Binds to the Cancer Drug Ruxolitinib with an Active Conformation
Plos One, 9, 2014
|
|
4LNP
 
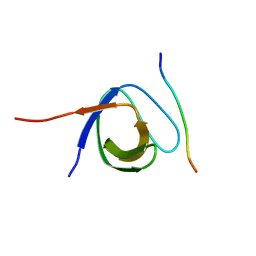 | | The first SH3 domain from CAP/Ponsin in complex with proline rich peptide from Vinculin | | Descriptor: | Sorbin and SH3 domain-containing protein 1, Vinculin | | Authors: | Zhao, D, Li, F, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2013-07-11 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
4LN2
 
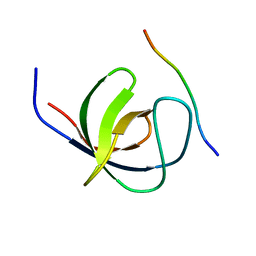 | | The second SH3 domain from CAP/Ponsin in complex with proline rich peptide from Vinculin | | Descriptor: | Sorbin and SH3 domain-containing protein 1, proline rich peptide | | Authors: | Zhao, D, Li, F, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2013-07-11 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
3DAP
 
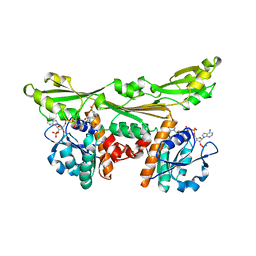 | | C. GLUTAMICUM DAP DEHYDROGENASE IN COMPLEX WITH NADP+ AND THE INHIBITOR 5S-ISOXAZOLINE | | Descriptor: | (2S,5',S)-2-AMINO-3-(3-CARBOXY-2-ISOXAZOLIN-5-YL)PROPANOIC ACID, DIAMINOPIMELIC ACID DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Scapin, G, Cirilli, M, Reddy, S.G, Gao, Y, Vederas, J.C, Blanchard, J.S. | | Deposit date: | 1997-12-29 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate and inhibitor binding sites in Corynebacterium glutamicum diaminopimelate dehydrogenase.
Biochemistry, 37, 1998
|
|
2IUH
 
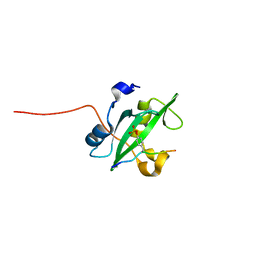 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain in complex with c-Kit phosphotyrosyl peptide | | Descriptor: | C-KIT PHOSPHOTYROSYL PEPTIDE, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY ALPHA SUBUNIT | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
1L3W
 
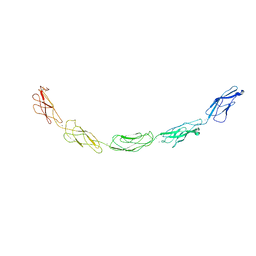 | | C-cadherin Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Boggon, T.J, Murray, J, Chappuis-Flament, S, Wong, E, Gumbiner, B.M, Shapiro, L. | | Deposit date: | 2002-03-01 | | Release date: | 2002-04-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | C-cadherin ectodomain structure and implications for cell adhesion mechanisms
Science, 296, 2002
|
|
4CU2
 
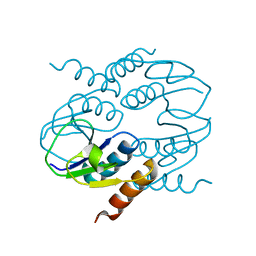 | | C-terminal domain of CTP1L endolysin mutant V195P that reduces autoproteolysis | | Descriptor: | ENDOLYSIN | | Authors: | Dunne, M, Mertens, H.D.T, Garefalaki, V, Jeffries, C.M, Thompson, A, Lemke, E.A, Svergun, D.I, Mayer, M.J, Narbad, A, Meijers, R. | | Deposit date: | 2014-03-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Cd27L and Ctp1L Endolysins Targeting Clostridia Contain a Built-in Trigger and Release Factor.
Plos Pathog., 10, 2014
|
|
4CU5
 
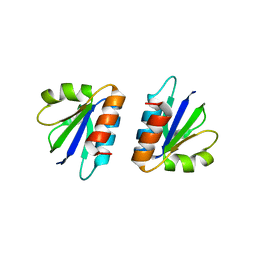 | | C-terminal domain of endolysin from phage CD27L is a trigger and release factor | | Descriptor: | ENDOLYSIN | | Authors: | Dunne, M, Mertens, H.D.T, Garefalaki, V, Jeffries, C.M, Thompson, A, Lemke, E.A, Svergun, D.I, Mayer, M.J, Narbad, A, Meijers, R. | | Deposit date: | 2014-03-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Cd27L and Ctp1L Endolysins Targeting Clostridia Contain a Built-in Trigger and Release Factor.
Plos Pathog., 10, 2014
|
|
1KTP
 
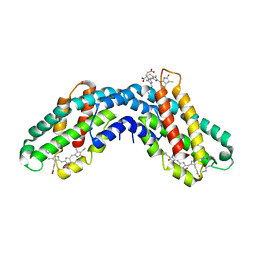 | |
6ELK
 
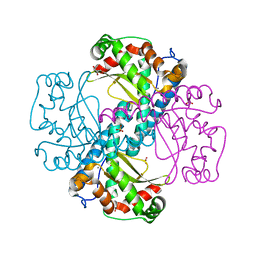 | | C.elegans MnSOD-3 mutant - Q142H | | Descriptor: | GLYCEROL, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hunter, G.J, Trinh, C.H, Hunter, T. | | Deposit date: | 2017-09-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Single Mutation is Sufficient to Modify the Metal Selectivity and Specificity of a Eukaryotic Manganese Superoxide Dismutase to Encompass Iron.
Chemistry, 24, 2018
|
|
6EY5
 
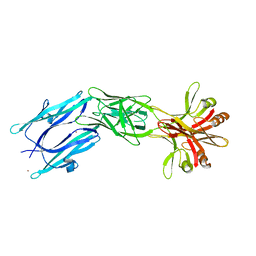 | | C-terminal part (residues 224-515) of PorM | | Descriptor: | T9SS component cytoplasmic membrane protein PorM, ZINC ION | | Authors: | Leone, P, Cambillau, C, Roussel, A. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space.
Nat Commun, 9, 2018
|
|
8H8A
 
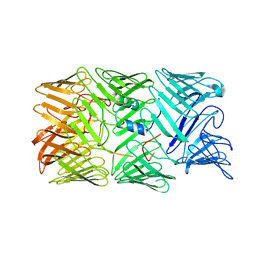 | | Type VI secretion system effector RhsP in its post-autoproteolysis and monomeric form | | Descriptor: | C-terminal peptide from Putative Rhs-family protein, Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
4XLG
 
 | | C. glabrata Slx1 in complex with Slx4CCD. | | Descriptor: | CHLORIDE ION, Structure-specific endonuclease subunit SLX1, Structure-specific endonuclease subunit SLX4, ... | | Authors: | Gaur, V, Wyatt, H.D.M, Komorowska, W, Szczepanowski, R.H, de Sanctis, D, Gorecka, K.M, West, S.C, Nowotny, M. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease.
Cell Rep, 10, 2015
|
|
4XM5
 
 | | C. glabrata Slx1. | | Descriptor: | CHLORIDE ION, Structure-specific endonuclease subunit SLX1, ZINC ION | | Authors: | Gaur, V, Wyatt, H.D.M, Komorowska, W, Szczepanowski, R.H, de Sanctis, D, Gorecka, K.M, West, S.C, Nowotny, M. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease.
Cell Rep, 10, 2015
|
|
8D4Y
 
 | | C-terminal SANT-SLIDE domain of human Chromodomain-helicase-DNA-binding protein 4 (CHD4) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Moghaddas Sani, H, Deshpande, C.N, Panjikar, S, Patel, K, Mackay, J.P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The role of auxiliary domains in modulating CHD4 activity suggests mechanistic commonality between enzyme families.
Nat Commun, 13, 2022
|
|
5HKN
 
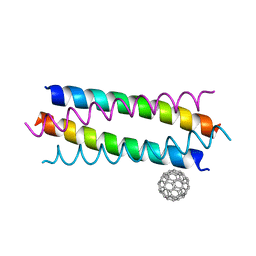 | | Crystal structure de novo designed fullerene organizing protein complex with fullerene | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | Authors: | Paul, J, Acharya, R, Kim, K.-H, Kim, Y.H, Kim, N.H, Grigoryan, G, DeGardo, W.F. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
