2R1V
 
 | |
2R1T
 
 | |
4O7K
 
 | | Crystal structure of Oncogenic Suppression Activity Protein - A Plasmid Fertility Inhibition Factor | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, PHOSPHATE ION, ... | | Authors: | Maindola, P, Goyal, P, Arulandu, A. | | Deposit date: | 2013-12-25 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Multiple enzymatic activities of ParB/Srx superfamily mediate sexual conflict among conjugative plasmids
Nat Commun, 5, 2014
|
|
2ETH
 
 | |
3PKI
 
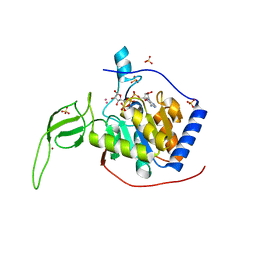 | | Human SIRT6 crystal structure in complex with ADP ribose | | Descriptor: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
2H24
 
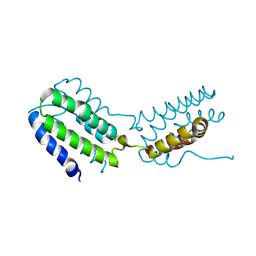 | | Crystal structure of human IL-10 | | Descriptor: | Interleukin-10 | | Authors: | Yoon, S.I, Walter, M.R. | | Deposit date: | 2006-05-18 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes mediate interleukin-10 receptor 2 (IL-10R2) binding to IL-10 and assembly of the signaling complex.
J.Biol.Chem., 281, 2006
|
|
3V6G
 
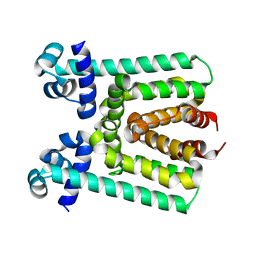 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
6YWY
 
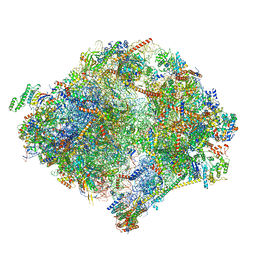 | | The structure of the mitoribosome from Neurospora crassa with bound tRNA at the P-site | | Descriptor: | 16S rRNA, 23S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
3CRP
 
 | | A heterospecific leucine zipper tetramer | | Descriptor: | GCN4 leucine zipper, SODIUM ION | | Authors: | Liu, J. | | Deposit date: | 2008-04-07 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A heterospecific leucine zipper tetramer.
Chem.Biol., 15, 2008
|
|
6NMA
 
 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
3V78
 
 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | ETHIDIUM, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
6YWX
 
 | | The structure of the mitoribosome from Neurospora crassa with tRNA bound to the E-site | | Descriptor: | 16S rRNA, 23S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
7WDC
 
 | | Crystal Structure of Cyanobacterial Circadian Clock Protein KaiC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Furuike, Y, Akiyama, S. | | Deposit date: | 2021-12-21 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Highly sensitive tryptophan fluorescence probe for detecting rhythmic conformational changes of KaiC in the cyanobacterial circadian clock system.
Biochem.J., 479, 2022
|
|
6NME
 
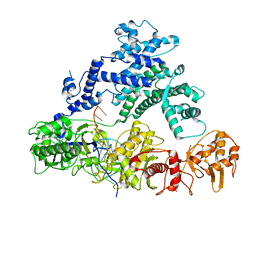 | | Structure of LbCas12a-crRNA | | Descriptor: | Cpf1, MAGNESIUM ION, crRNA | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.67 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NJT
 
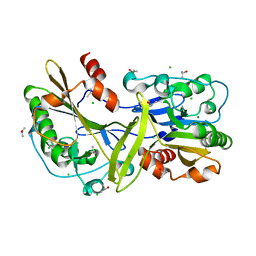 | | Mouse endonuclease G mutant - H97A | | Descriptor: | CHLORIDE ION, Endonuclease G, mitochondrial, ... | | Authors: | Vander Zanden, C.M, Ho, E.N, Czarny, R.S, Robertson, A.B, Ho, P.S. | | Deposit date: | 2019-01-04 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural adaptation of vertebrate endonuclease G for 5-hydroxymethylcytosine recognition and function.
Nucleic Acids Res., 48, 2020
|
|
6YWE
 
 | | The structure of the mitoribosome from Neurospora crassa in the P/E tRNA bound state | | Descriptor: | 16S rRNA, 23S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6NM9
 
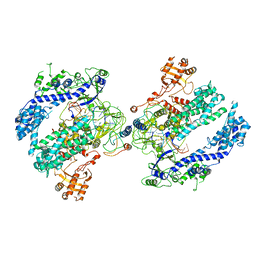 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4 dimer | | Descriptor: | AcrVA4, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6YWV
 
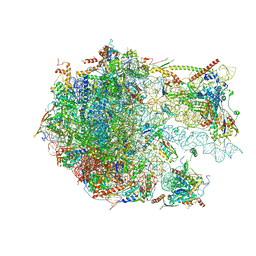 | | The structure of the Atp25 bound assembly intermediate of the mitoribosome from Neurospora crassa | | Descriptor: | 23 S rRNA, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
7ZUB
 
 | | Cryo-EM structure of the indirubin-bound Hsp90-XAP2-AHR complex | | Descriptor: | (3~{Z})-3-(3-oxidanylidene-1~{H}-indol-2-ylidene)-1~{H}-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, ... | | Authors: | Gruszczyk, J, Savva, C.G, Lai-Kee-Him, J, Bous, J, Ancelin, A, Kwong, H.S, Grandvuillemin, L, Bourguet, W. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the agonist-bound Hsp90-XAP2-AHR cytosolic complex.
Nat Commun, 13, 2022
|
|
2YRP
 
 | | Solution structure of the TIG domain from Human Nuclear factor of activated T-cells, cytoplasmic 4 | | Descriptor: | Nuclear factor of activated T-cells, cytoplasmic 4 | | Authors: | Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TIG domain from Human Nuclear factor of activated T-cells, cytoplasmic 4
To be Published
|
|
3O0G
 
 | | Crystal Structure of Cdk5:p25 in complex with an ATP analogue | | Descriptor: | Cell division protein kinase 5, Cyclin-dependent kinase 5 activator 1, {4-amino-2-[(4-chlorophenyl)amino]-1,3-thiazol-5-yl}(3-nitrophenyl)methanone | | Authors: | Mapelli, M. | | Deposit date: | 2010-07-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Defining Cdk5 ligand chemical space with small molecule inhibitors of Tau phosphorylation
Chem.Biol., 12, 2005
|
|
1EY1
 
 | | SOLUTION STRUCTURE OF ESCHERICHIA COLI NUSB | | Descriptor: | ANTITERMINATION FACTOR NUSB | | Authors: | Altieri, A.S, Mazzulla, M.J, Horita, D.A, Coats, R.H, Wingfield, P.T, Byrd, R.A. | | Deposit date: | 2000-05-05 | | Release date: | 2000-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the transcriptional antiterminator NusB from Escherichia coli.
Nat.Struct.Biol., 7, 2000
|
|
6HL0
 
 | | Crystal Structure of Farnesoid X receptor (FXR) with bound NCoA-2 peptide | | Descriptor: | Bile acid receptor, NCoA-2 peptide (Nuclear receptor coactivator 2), LYS-GLU-ASN-ALA-LEU-LEU-ARG-TYR-LEU-LEU-ASP-LYS-ASP | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular tuning of farnesoid X receptor partial agonism.
Nat Commun, 10, 2019
|
|
1U6G
 
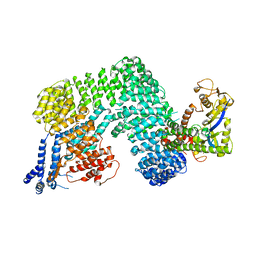 | | Crystal Structure of The Cand1-Cul1-Roc1 Complex | | Descriptor: | Cullin homolog 1, RING-box protein 1, TIP120 protein, ... | | Authors: | Goldenberg, S.J, Shumway, S.D, Cascio, T.C, Garbutt, K.C, Liu, J, Xiong, Y, Zheng, N. | | Deposit date: | 2004-07-29 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases
Cell(Cambridge,Mass.), 119, 2004
|
|
2ZTA
 
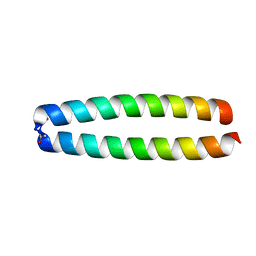 | | X-RAY STRUCTURE OF THE GCN4 LEUCINE ZIPPER, A TWO-STRANDED, PARALLEL COILED COIL | | Descriptor: | GCN4 LEUCINE ZIPPER | | Authors: | O'Shea, E.K, Klemm, J.D, Kim, P.S, Alber, T. | | Deposit date: | 1991-07-05 | | Release date: | 1992-10-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil.
Science, 254, 1991
|
|
