1G5W
 
 | | SOLUTION STRUCTURE OF HUMAN HEART-TYPE FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID-BINDING PROTEIN | | Authors: | Luecke, C, Rademacher, M, Zimmerman, A, van Moerkerk, H.T.B, Veerkamp, J.H, Rueterjans, H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Spin-system heterogeneities indicate a selected-fit mechanism in fatty acid binding to heart-type fatty acid-binding protein (H-FABP).
Biochem.J., 354, 2001
|
|
4YYE
 
 | |
8OO1
 
 | |
8SLY
 
 | | Rat TRPV2 bound with 2 CBD ligands in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 2, ... | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cannabidiol sensitizes TRPV2 channels to activation by 2-APB.
Elife, 12, 2023
|
|
3JQK
 
 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 (H32 FORM) | | Descriptor: | ACETATE ION, Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7T4Z
 
 | | Crystal structure of the molybdate-binding periplasmic protein ModA from the bacteria Pseudomonsa aeruginosa in ligand-free form | | Descriptor: | AMMONIUM ION, GLYCEROL, Molybdate-binding periplasmic protein, ... | | Authors: | Ngu, D.H.Y, Luo, Z, Lim, B.Y.J, Kobe, B. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Impact of Chromate on Pseudomonas aeruginosa Molybdenum Homeostasis.
Front Microbiol, 13, 2022
|
|
2BEF
 
 | | CRYSTAL STRUCTURE OF NDP KINASE COMPLEXED WITH MG, ADP, AND BEF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Xu, Y.W, Cherfils, J. | | Deposit date: | 1998-05-26 | | Release date: | 1998-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AlF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2WOV
 
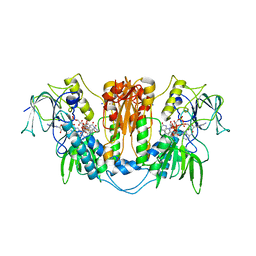 | | Trypanosoma brucei trypanothione reductase with bound NADP. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Alphey, M.S, Fairlamb, A.H. | | Deposit date: | 2009-07-30 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2GA3
 
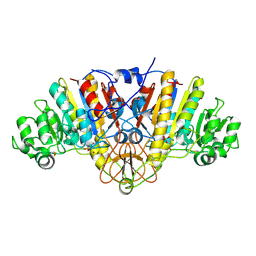 | |
2WPF
 
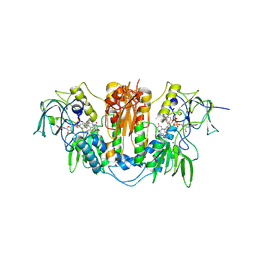 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00085762) | | Descriptor: | 3-[(4S)-6-CHLORO-2-METHYL-4-(4-METHYLPHENYL)QUINAZOLIN-3(4H)-YL]-N,N-DIMETHYLPROPAN-1-AMINE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-06 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
4H8I
 
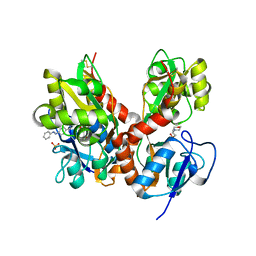 | | Structure of GluK2-LBD in complex with GluAzo | | Descriptor: | (4R)-4-[(2E)-3-{4-[(E)-phenyldiazenyl]phenyl}prop-2-en-1-yl]-L-glutamic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Reiter, A, Skerra, A, Trauner, D, Schiefner, A. | | Deposit date: | 2012-09-22 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A photoswitchable neurotransmitter analogue bound to its receptor.
Biochemistry, 52, 2013
|
|
2WOW
 
 | | Trypanosoma brucei trypanothione reductase with NADP and trypanothione bound | | Descriptor: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alphey, M.S, Fairlamb, A.H. | | Deposit date: | 2009-07-30 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
4I3Q
 
 | |
3D0U
 
 | | Crystal Structure of Lysine Riboswitch Bound to Lysine | | Descriptor: | IRIDIUM HEXAMMINE ION, LYSINE, Lysine Riboswitch RNA | | Authors: | Garst, A.D, Heroux, A, Rambo, R.P, Batey, R.T. | | Deposit date: | 2008-05-02 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the lysine riboswitch regulatory mRNA element.
J.Biol.Chem., 283, 2008
|
|
4LWT
 
 | | The 1.6A Crystal Structure of Humanized Xenopus MDM2 with RO5027344 | | Descriptor: | (3S)-3-[(3R)-1-acetylpiperidin-3-yl]-6-chloro-3-(3-chlorobenzyl)-1,3-dihydro-2H-indol-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Kammlott, U. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent and selective spiroindolinone MDM2 inhibitor, RO8994, for cancer therapy.
Bioorg.Med.Chem., 22, 2014
|
|
9BOU
 
 | |
8SO2
 
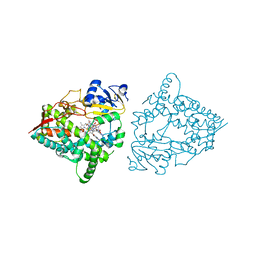 | | Human CYP3A4 bound to a substrate | | Descriptor: | CAFFEINE, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Interaction of CYP3A4 with caffeine: First insights into multiple substrate binding.
J.Biol.Chem., 299, 2023
|
|
4KR2
 
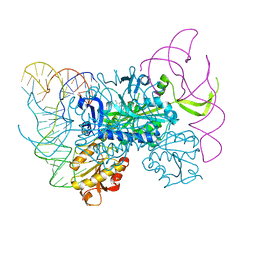 | | Glycyl-tRNA synthetase in complex with tRNA-Gly | | Descriptor: | ADENOSINE MONOPHOSPHATE, Gly-tRNA-CCC, Glycine--tRNA ligase | | Authors: | Qin, X, Hao, Z, Tian, Q, Zhang, Z, Zhou, C, Xie, W. | | Deposit date: | 2013-05-16 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.292 Å) | | Cite: | Cocrystal Structures of Glycyl-tRNA Synthetase in Complex with tRNA Suggest Multiple Conformational States in Glycylation
J.Biol.Chem., 289, 2014
|
|
4KR3
 
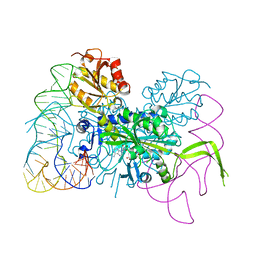 | | Glycyl-tRNA synthetase mutant E71G in complex with tRNA-Gly | | Descriptor: | GLYCINE, Gly-tRNA-CCC, Glycine--tRNA ligase, ... | | Authors: | Qin, X, Hao, Z, Tian, Q, Zhang, Z, Zhou, C, Xie, W. | | Deposit date: | 2013-05-16 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.235 Å) | | Cite: | Cocrystal Structures of Glycyl-tRNA Synthetase in Complex with tRNA Suggest Multiple Conformational States in Glycylation
J.Biol.Chem., 289, 2014
|
|
8SO1
 
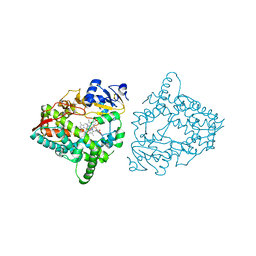 | | Human CYP3A4 bound to three caffeine molecules | | Descriptor: | CAFFEINE, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Interaction of CYP3A4 with caffeine: First insights into multiple substrate binding.
J.Biol.Chem., 299, 2023
|
|
1BXA
 
 | | AMICYANIN REDUCED, PH 4.4, 1.3 ANGSTROMS | | Descriptor: | COPPER (I) ION, PROTEIN (AMICYANIN) | | Authors: | Cunane, L.M, Chen, Z.W, Durley, R.C.E, Mathews, F.S. | | Deposit date: | 1998-10-01 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for interprotein complex-dependent effects on the redox properties of amicyanin.
Biochemistry, 37, 1998
|
|
4K5Z
 
 | | Crystal Structure of Human Chymase in Complex with Fragment Inhibitor 6-chloro-2,3-dihydro-1H-isoindol-1-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-2,3-dihydro-1H-isoindol-1-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
1TLX
 
 | | THERMOLYSIN (NATIVE) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, LYSINE, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-11-02 | | Release date: | 2000-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
7CBI
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)pentyl (2~{S},3~{R})-2-azanyl-3-oxidanyl-butanoate, GLYCEROL, ... | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2020-06-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
7U8F
 
 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA damage-binding protein 1, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
