3CWG
 
 | | Unphosphorylated mouse STAT3 core fragment | | Descriptor: | Signal transducer and activator of transcription 3 | | Authors: | Ren, Z, Mao, X, Mertens, C, Krishnaraj, R, Qin, J, Mandal, P.K, Romanowshi, M.J, McMurray, J.S. | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of unphosphorylated STAT3 core fragment.
Biochem.Biophys.Res.Commun., 374, 2008
|
|
2X74
 
 | | Human foamy virus integrase - catalytic core. | | Descriptor: | INTEGRASE | | Authors: | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Legrand, P, Silhan, J, Lewit-Bentley, A. | | Deposit date: | 2010-02-23 | | Release date: | 2010-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6GUB
 
 | | CDK2/CyclinA in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6P5K
 
 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 3) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
5TW3
 
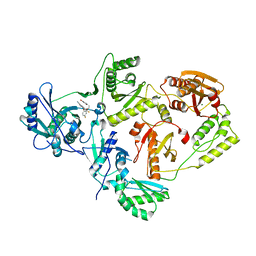 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2016-11-11 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural and Preclinical Studies of Computationally Designed Non-Nucleoside Reverse Transcriptase Inhibitors for Treating HIV infection.
Mol. Pharmacol., 91, 2017
|
|
8B6H
 
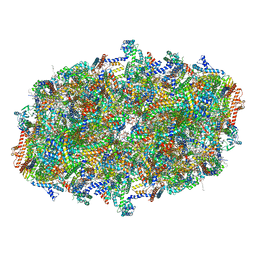 | | Cryo-EM structure of cytochrome c oxidase dimer (complex IV) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8B7Q
 
 | | Cryo-EM structure for the mouse LEPR-CRH2:Leptin:LEPR-Ig complex following symmetry expansion in combination with local refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-09-30 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2C2V
 
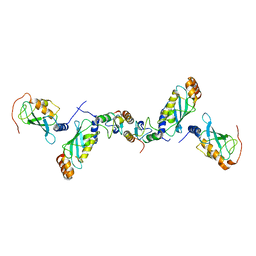 | | Crystal structure of the CHIP-UBC13-UEV1a complex | | Descriptor: | STIP1 homology and U box-containing protein 1, Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 1 | | Authors: | Zhang, M, Roe, S.M, Pearl, L.H. | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex.
Mol. Cell, 20, 2005
|
|
6ZVK
 
 | | The Halastavi arva virus (HalV) intergenic region IRES promotes translation by the simplest possible initiation mechanism | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES17, ... | | Authors: | Abaeva, I.S, Vicens, Q, Bochler, A, Soufari, H, Simonetti, A, Pestova, T, Hashem, Y, Hellen, C.U.T. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The Halastavi arva Virus Intergenic Region IRES Promotes Translation by the Simplest Possible Initiation Mechanism.
Cell Rep, 33, 2020
|
|
6INL
 
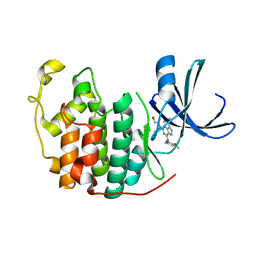 | | Crystal structure of CDK2 IN complex with Inhibitor CVT-313 | | Descriptor: | 2,2'-{[6-{[(4-methoxyphenyl)methyl]amino}-9-(propan-2-yl)-9H-purin-2-yl]azanediyl}di(ethan-1-ol), Cyclin-dependent kinase 2 | | Authors: | Talapati, S.R, Krishnamurthy, N.R. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of cyclin-dependent kinase 2 (CDK2) in complex with the specific and potent inhibitor CVT-313.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7V7L
 
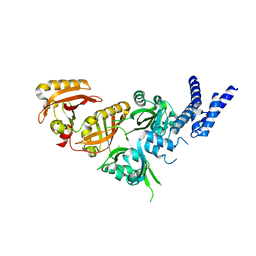 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
7VOI
 
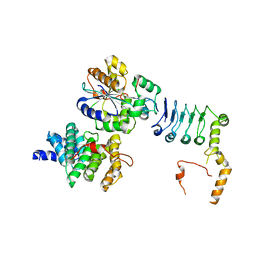 | | Structure of the human CNOT1(MIF4G)-CNOT6L-CNOT7 complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 6-like, CCR4-NOT transcription complex subunit 7 | | Authors: | Bartlam, M, Zhang, Q. | | Deposit date: | 2021-10-13 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.38 Å) | | Cite: | Structure of the human Ccr4-Not nuclease module using X-ray crystallography and electron paramagnetic resonance spectroscopy distance measurements.
Protein Sci., 31, 2022
|
|
7V7W
 
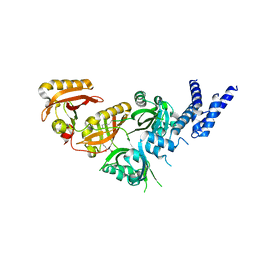 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex with oleoylethanolamide (OEA) | | Descriptor: | (Z)-N-(2-hydroxyethyl)octadec-9-enamide, Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Zhang, M, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
7ZTL
 
 | |
7MQ9
 
 | | Cryo-EM structure of the human SSU processome, state pre-A1* | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Vanden Broeck, A, Singh, S, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
7MQ8
 
 | | Cryo-EM structure of the human SSU processome, state pre-A1 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Vanden Broeck, A, Singh, S, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
7NAC
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Composite model | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6GUC
 
 | | CDK2/CyclinA in complex with SU9516 | | Descriptor: | (3Z)-3-(1H-IMIDAZOL-5-YLMETHYLENE)-5-METHOXY-1H-INDOL-2(3H)-ONE, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GUK
 
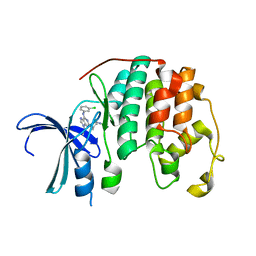 | | CDK2 in complex with CGP74514A | | Descriptor: | Cyclin-dependent kinase 2, ~{N}2-[(1~{R},2~{S})-2-azanylcyclohexyl]-~{N}6-(3-chlorophenyl)-9-ethyl-purine-2,6-diamine | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
3KOR
 
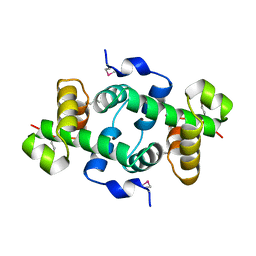 | | Crystal structure of a putative Trp repressor from Staphylococcus aureus | | Descriptor: | Possible Trp repressor | | Authors: | Lam, R, Vodsedalek, J, Lam, K, Romanov, V, Battaile, K.P, Beletskaya, I, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-11-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a putative Trp repressor from Staphylococcus aureus
To be Published
|
|
8CHF
 
 | | cryo-EM Structure of Craf:14-3-3:Mek1 | | Descriptor: | 14-3-3 protein zeta isoform X1, 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Dedden, D, Ulrich, G. | | Deposit date: | 2023-02-07 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM Structures of CRAF 2 /14-3-3 2 and CRAF 2 /14-3-3 2 /MEK1 2 Complexes.
J.Mol.Biol., 436, 2024
|
|
8OHN
 
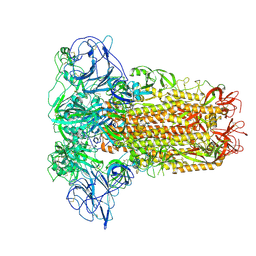 | | Human Coronavirus HKU1 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pronker, M.F, Hurdiss, D.L. | | Deposit date: | 2023-03-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8OPN
 
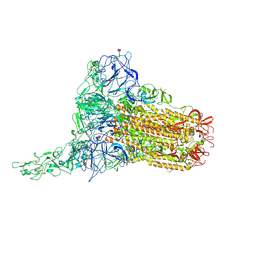 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8OPO
 
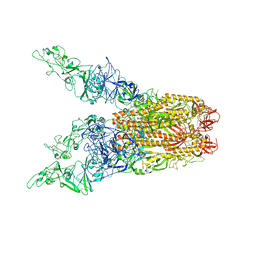 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (3-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8OPM
 
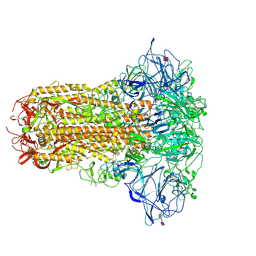 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
