3IT9
 
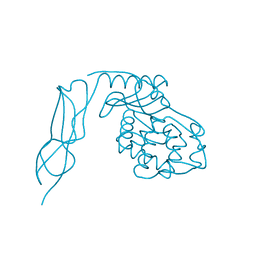 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in apo state | | Descriptor: | D-alanyl-D-alanine carboxypeptidase dacC, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
5IEE
 
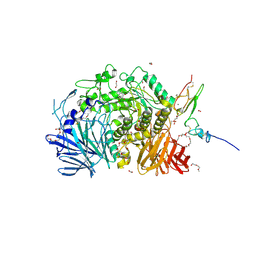 | | Murine endoplasmic reticulum alpha-glucosidase II with 1-deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3WQ1
 
 | |
4X32
 
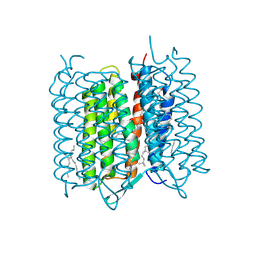 | | Bacteriorhodopsin ground state structure collected in cryo conditions from crystals obtained in LCP with PEG as a precipitant. | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, Standfuss, J. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lipidic cubic phase serial millisecond crystallography using synchrotron radiation.
Iucrj, 2, 2015
|
|
5L2J
 
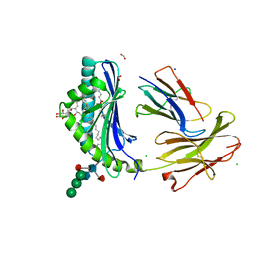 | | Crystal Structure of human CD1b in complex with C36-GMM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-[(2R,3R)-3-hydroxy-2-tetradecyldocosanoyl]-alpha-L-idopyranose, ACETATE ION, ... | | Authors: | Gras, S, Shahine, A, Le Nours, J, Rossjohn, J. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | T cell receptor recognition of CD1b presenting a mycobacterial glycolipid.
Nat Commun, 7, 2016
|
|
1C04
 
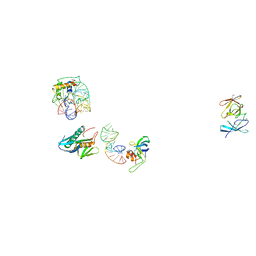 | | IDENTIFICATION OF KNOWN PROTEIN AND RNA STRUCTURES IN A 5 A MAP OF THE LARGE RIBOSOMAL SUBUNIT FROM HALOARCULA MARISMORTUI | | Descriptor: | 23S RRNA FRAGMENT, RIBOSOMAL PROTEIN L11, RIBOSOMAL PROTEIN L14, ... | | Authors: | Ban, N, Nissen, P, Capel, M, Moore, P.B, Steitz, T.A. | | Deposit date: | 1999-07-14 | | Release date: | 1999-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Placement of protein and RNA structures into a 5 A-resolution map of the 50S ribosomal subunit.
Nature, 400, 1999
|
|
6SEB
 
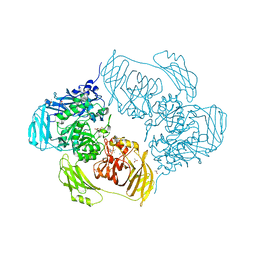 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, ACETATE ION, Beta-galactosidase, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Kaminska, P, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6LWU
 
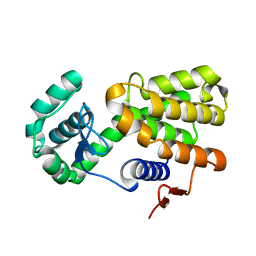 | | Complex of Gynuella sunshinyii GH46 chitosanase GsCsn46A E19A with chitopentaose | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, chitosanase | | Authors: | Qin, Z, Wang, Y. | | Deposit date: | 2020-02-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.419 Å) | | Cite: | Complex of Gynuella sunshinyii GH46 chitosanase GsCsn46A E19A with chitopentaose
To Be Published
|
|
4X31
 
 | | Room temperature structure of bacteriorhodopsin from lipidic cubic phase obtained with serial millisecond crystallography using synchrotron radiation | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, James, D, Wang, D, White, T, Zatsepin, N, Shilova, A, Nelson, G, Liu, H, Johansson, L, Heymann, M, Jaeger, K, Metz, M, Wickstrand, C, Wu, W, Baath, P, Berntsen, P, Oberthuer, D, Panneels, V, Cherezov, V, Chapman, H, Spence, J, Schertler, G, Neutze, R, Moraes, I, Burghammer, M, Standfuss, J, Weierstall, U. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipidic cubic phase serial millisecond crystallography using synchrotron radiation.
Iucrj, 2, 2015
|
|
5L2K
 
 | | Crystal structure of GEM42 TCR-CD1b-GMM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-[(2R,3R)-3-hydroxy-2-tetradecyldocosanoyl]-alpha-L-idopyranose, Beta-2-microglobulin, ... | | Authors: | Gras, S, Shahine, A, Le Nours, J, Rossjohn, J. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | T cell receptor recognition of CD1b presenting a mycobacterial glycolipid.
Nat Commun, 7, 2016
|
|
6LYN
 
 | | CD146 D4-D5/AA98 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AA98 Fab heavy chain, ... | | Authors: | Chen, X, Yan, X. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.776 Å) | | Cite: | Structure basis for AA98 inhibition on the activation of endothelial cells mediated by CD146.
Iscience, 24, 2021
|
|
7B1O
 
 | | Crystal structure of the indoleamine 2,3-dioxygenase 1 (IDO1) in complex with compound 22 | | Descriptor: | 4-chloranyl-N-[(1R)-1-[(1S,5R)-3-quinolin-4-yloxy-6-bicyclo[3.1.0]hexanyl]propyl]benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lammens, A, Krapp, S, Lewis, R.T, Hamilton, M.M. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
3IN9
 
 | | Crystal structure of heparin lyase I complexed with disaccharide heparin | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparin lyase I | | Authors: | Han, Y.H, Ryu, K.S, Kim, H.Y, Jeon, Y.H. | | Deposit date: | 2009-08-12 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of heparin depolymerization by heparin lyase I
J.Biol.Chem., 284, 2009
|
|
1C40
 
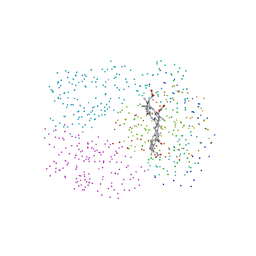 | | BAR-HEADED GOOSE HEMOGLOBIN (AQUOMET FORM) | | Descriptor: | PROTEIN (HEMOGLOBIN (ALPHA CHAIN)), PROTEIN (HEMOGLOBIN (BETA CHAIN)), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, S, Liu, X, Jing, H, Hua, Z, Zhang, R, Lu, G. | | Deposit date: | 1999-08-03 | | Release date: | 1999-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Avian haemoglobins and structural basis of high affinity for oxygen: structure of bar-headed goose aquomet haemoglobin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6M6T
 
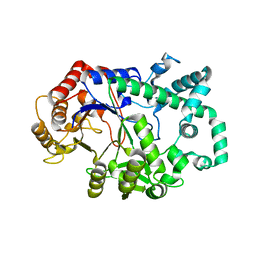 | | Amylomaltase from Streptococcus agalactiae in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-1,5-anhydro-D-glucitol, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Wangkanont, K, Tumhom, S, Pongsawasdi, P. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Streptococcus agalactiae amylomaltase offers insight into the transglycosylation mechanism and the molecular basis of thermostability among amylomaltases.
Sci Rep, 11, 2021
|
|
8T6V
 
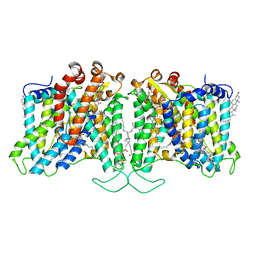 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatostilbene-2,2'-Disulfonic Acid (DIDS) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | Authors: | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2023-06-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6P5O
 
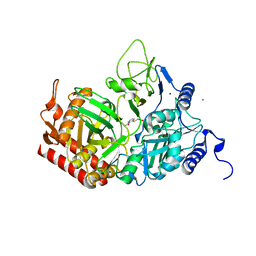 | |
6SNY
 
 | | Synthetic mimic of an EPCR-binding PfEMP1 bound to EPCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endothelial protein C receptor, ... | | Authors: | Barber, N.M, Higgins, M.K. | | Deposit date: | 2019-08-27 | | Release date: | 2019-09-04 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure-Guided Design of a Synthetic Mimic of an Endothelial Protein C Receptor-Binding PfEMP1 Protein.
Msphere, 6, 2021
|
|
6SU2
 
 | |
9CYL
 
 | | Structure of LAG3 loop1 deletion bound to the MHC class II molecule I-A(b) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Class-II-associated invariant chain peptide, H-2 class II histocompatibility antigen, ... | | Authors: | Ming, Q, Tran, T.H, Antfolk, D, Luca, V.C. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (4.66 Å) | | Cite: | Structural basis for LAG3 interactions with MHC class II
To Be Published
|
|
9CYM
 
 | | Structure of LAG3 bound to the MHC class II molecule I-A(b) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Class-II-associated invariant chain peptide, H-2 class II histocompatibility antigen, ... | | Authors: | Ming, Q, Antfolk, D, Tran, T.H, Luca, V.C. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Structural basis for LAG3 interactions with MHC class II
To Be Published
|
|
6ATK
 
 | |
5LGV
 
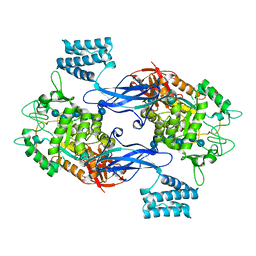 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltooctaose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Mia, F, Barclay, J.E, Tang, M, Gorelik, A, Rashid, A.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
8T6U
 
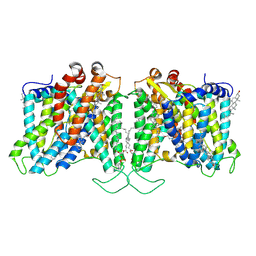 | | Cryo-EM structure of human Anion Exchanger 1 bound to Dipyridamole | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2023-06-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3IGU
 
 | | Crystal structure of human alpha-N-acetylgalactosaminidase, covalent intermediate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clark, N.E, Garman, S.C. | | Deposit date: | 2009-07-28 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The 1.9 a structure of human alpha-N-acetylgalactosaminidase: The molecular basis of Schindler and Kanzaki diseases
J.Mol.Biol., 393, 2009
|
|
