4PEK
 
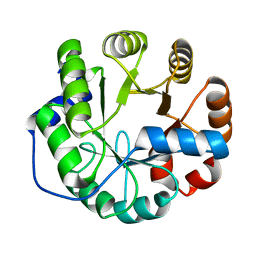 | | Crystal structure of a computationally designed retro-aldolase, RA114.3 | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
4PEJ
 
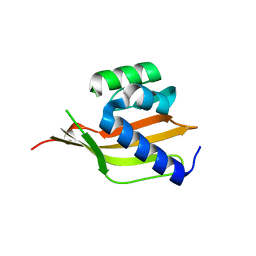 | | Crystal structure of a computationally designed retro-aldolase, RA110.4 (Cys free) | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
8J98
 
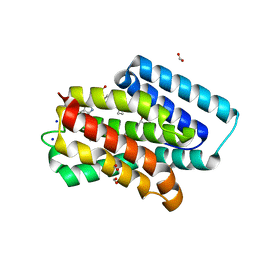 | | A near-infrared fluorescent protein of de novo backbone design | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Hu, X, Xu, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | De novo backbone design for monomerization of near-infrared fluorescent protein
To Be Published
|
|
5YXI
 
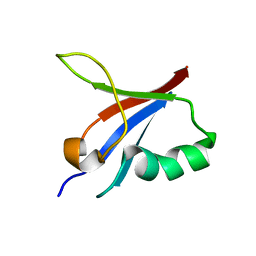 | | Designed protein dRafX6 | | Descriptor: | Designed protein dRafX6 | | Authors: | Liu, R. | | Deposit date: | 2017-12-05 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo sequence redesign of a functional Ras-binding domain globally inverted the surface charge distribution and led to extreme thermostability.
Biotechnol.Bioeng., 118, 2021
|
|
7JRQ
 
 | | Crystallographically Characterized De Novo Designed Mn-Diphenylporphyrin Binding Protein | | Descriptor: | CHLORIDE ION, GLYCEROL, MPP1, ... | | Authors: | Mann, S.I, DeGrado, W.F. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De Novo Design, Solution Characterization, and Crystallographic Structure of an Abiological Mn-Porphyrin-Binding Protein Capable of Stabilizing a Mn(V) Species.
J.Am.Chem.Soc., 143, 2021
|
|
7XAD
 
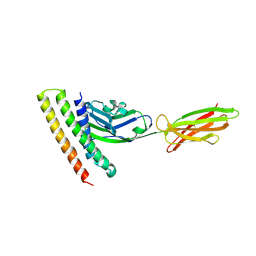 | | Crystal strucutre of PD-L1 and DBL2_02 designed protein binder | | Descriptor: | DBL2_02 binder, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7XYQ
 
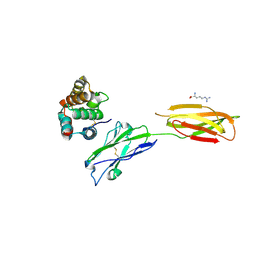 | | Crystal strucutre of PD-L1 and the computationally designed DBL1_03 protein binder | | Descriptor: | ARGININE, CD274 molecule, DBL1_03 | | Authors: | Liu, K, Xu, Z, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S. | | Deposit date: | 2022-06-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
4AF0
 
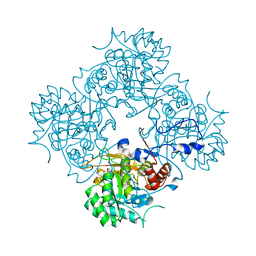 | | Crystal structure of cryptococcal inosine monophosphate dehydrogenase | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, MYCOPHENOLIC ACID, ... | | Authors: | Valkov, E, Stamp, A, Morrow, C.A, Kobe, B, Fraser, J.A. | | Deposit date: | 2012-01-15 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De Novo GTP Biosynthesis is Critical for Virulence of the Fungal Pathogen Cryptococcus Neoformans
Plos Pathog., 8, 2012
|
|
8BFE
 
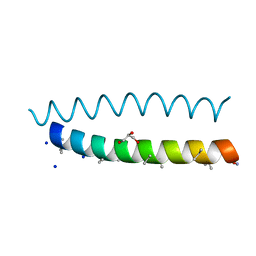 | | A dimeric de novo coiled-coil assembly: PK-2 (CC-TypeN-LaUbUcLd) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CC-TypeN-LaUbUcLd, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
7K3H
 
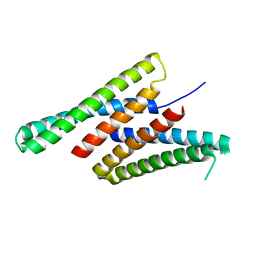 | | Crystal structure of deep network hallucinated protein 0217 | | Descriptor: | Network hallucinated protein 0217 | | Authors: | Pellock, S.J, Anishchenko, I, Chidyausiku, T.M, Bera, A.K, DiMaio, F, Baker, D. | | Deposit date: | 2020-09-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo protein design by deep network hallucination.
Nature, 600, 2021
|
|
8W0Q
 
 | | Pembrolizumab CDR-H3 Loop Mimic | | Descriptor: | Pembrolizumab CDR-H3 Loop Mimic | | Authors: | Feig, M, Roche, S.P. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | De Novo Synthesis and Structural Elucidation of CDR-H3 Loop Mimics.
Acs Chem.Biol., 2024
|
|
7JZL
 
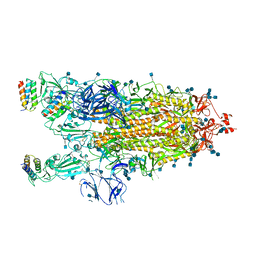 | |
7JZN
 
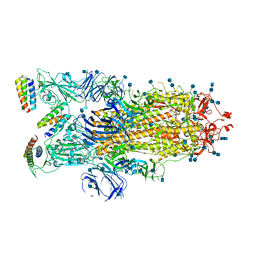 | |
7JZM
 
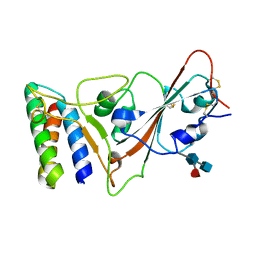 | |
7JZU
 
 | |
5A6N
 
 | | Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with compound 2 | | Descriptor: | 5-(3-SULFAMOYLPHENYL)-1H-1,2,3,4-TETRAZOL-1-IDE, DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, ... | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7KL9
 
 | |
5A6O
 
 | | Crystal structure of the apo form of the unphosphorylated human death associated protein kinase 3 (DAPK3) | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, S-1,2-PROPANEDIOL | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1O6J
 
 | | Tryparedoxin II from C.fasciculata solved by sulphur phasing | | Descriptor: | TRYPAREDOXIN II | | Authors: | Leonard, G.A, Micossi, E, Hunter, W.N. | | Deposit date: | 2002-10-04 | | Release date: | 2002-11-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | De Novo Phasing of Two Crystal Forms of Tryparedoxin II Using the Anomalous Scattering from S Atoms: A Combination of Small Signal and Medium Resolution Reveals This to be a General Tool for Solving Protein Crystal Structures
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1O81
 
 | | Tryparedoxin II from C.fasciculata solved by sulphur phasing | | Descriptor: | SULFATE ION, TRYPAREDOXIN II | | Authors: | Leonard, G.A, Micossi, E, Hunter, W.N. | | Deposit date: | 2002-11-21 | | Release date: | 2002-12-19 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | De Novo Phasing of Two Crystal Forms of Tryparedoxin II Using the Anomalous Scattering from S Atoms: A Combination of Small Signal and Medium Resolution Reveals This to be a General Tool for Solving Protein Crystal Structures
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1Z70
 
 | | 1.15A resolution structure of the formylglycine generating enzyme FGE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-alpha-formyglycine-generating enzyme, ... | | Authors: | Rudolph, M.G. | | Deposit date: | 2005-03-23 | | Release date: | 2005-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | De novo calcium/sulfur SAD phasing of the human formylglycine-generating enzyme using in-house data.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7XZR
 
 | | Crystal structure of TNIK-AMPPNP-thiopeptide TP15 complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Hamada, K, Vinogradov, A.A, Zhang, Y, Chang, J.S, Nishimura, H, Goto, Y, Onaka, H, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
J.Am.Chem.Soc., 144, 2022
|
|
7XZQ
 
 | | Crystal structure of TNIK-thiopeptide TP1 complex | | Descriptor: | 1,4-BUTANEDIOL, TRAF2 and NCK-interacting protein kinase, thiopeptide TP1 | | Authors: | Hamada, K, Vinogradov, A.A, Zhang, Y, Chang, J.S, Nishimura, H, Goto, Y, Onaka, H, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
J.Am.Chem.Soc., 144, 2022
|
|
3C9V
 
 | | C7 Symmetrized Structure of Unliganded GroEL at 4.7 Angstrom Resolution from CryoEM | | Descriptor: | 60 kDa chaperonin | | Authors: | Ludtke, S.J, Baker, M.L, Chen, D.H, Song, J.L, Chuang, D, Chiu, W. | | Deposit date: | 2008-02-18 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | De Novo Backbone Trace of GroEL from Single Particle Electron Cryomicroscopy.
Structure, 16, 2008
|
|
3CAU
 
 | | D7 symmetrized structure of unliganded GroEL at 4.2 Angstrom resolution by cryoEM | | Descriptor: | 60 kDa chaperonin | | Authors: | Ludtke, S.J, Baker, M.L, Chen, D.H, Song, J.L, Chuang, D, Chiu, W. | | Deposit date: | 2008-02-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | De Novo Backbone Trace of GroEL from Single Particle Electron Cryomicroscopy.
Structure, 16, 2008
|
|
