1S68
 
 | | Structure and Mechanism of RNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA Ligase 2 | | Authors: | Ho, C.K, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of RNA ligase.
Structure, 12, 2004
|
|
2HVS
 
 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 2'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2HVR
 
 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 3'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
3BA9
 
 | | Structural Basis for Inhbition of NAD-Dependent Ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, GLYCEROL, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Inhbition of NAD-Dependent Ligase
To be Published
|
|
6U75
 
 | |
2ZKF
 
 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DAP*DTP*DCP*(5CM)P*DGP*DGP*DTP*DGP*DA)-3'), DNA (5'-D(P*DCP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DAP*DGP*DA)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
2ZKE
 
 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DGP*DC)-3'), DNA (5'-D(*DGP*DCP*DAP*DAP*DTP*DCP*(5CM)P*DGP*DGP*DTP*DAP*DG)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
2C5U
 
 | | T4 RNA Ligase (Rnl1) Crystal Structure | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | El Omari, K, Ren, J, Bird, L.E, Bona, M.K, Klarmann, G, LeGrice, S.F.J, Stammers, D.K. | | Deposit date: | 2005-11-01 | | Release date: | 2005-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Molecular Architecture and Ligand Recognition Determinants for T4 RNA Ligase
J.Biol.Chem., 281, 2006
|
|
5COT
 
 | |
5COU
 
 | |
6WID
 
 | | Nucleotide incorporation intermediate into quaternary complex of human Polymerase Mu on a complementary DNA double-strand break substrate | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaminski, A.M, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2020-04-09 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of human DNA polymerase mu engaged on a DNA double-strand break.
Nat Commun, 11, 2020
|
|
6WIC
 
 | | Pre-catalytic quaternary complex of human Polymerase Mu on a complementary DNA double-strand break substrate | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaminski, A.M, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2020-04-09 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural snapshots of human DNA polymerase mu engaged on a DNA double-strand break.
Nat Commun, 11, 2020
|
|
5TT6
 
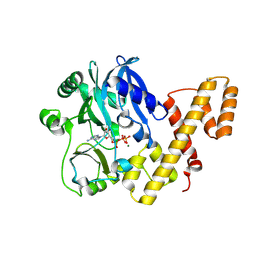 | | T4 RNA Ligase 1 (K99M) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, T4 RNA ligase 1 | | Authors: | Goldgur, Y, Unciuleac, M.-C, Shuman, S.H. | | Deposit date: | 2016-11-01 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD(+)-dependent polynucleotide ligases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6WIE
 
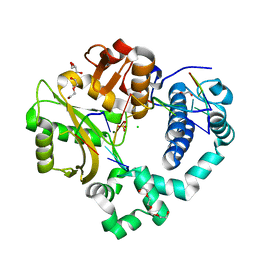 | | Post-catalytic nicked complex of human Polymerase Mu on a complementary DNA double-strand break substrate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*CP*G)-3'), ... | | Authors: | Kaminski, A.M, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2020-04-09 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of human DNA polymerase mu engaged on a DNA double-strand break.
Nat Commun, 11, 2020
|
|
7OTM
 
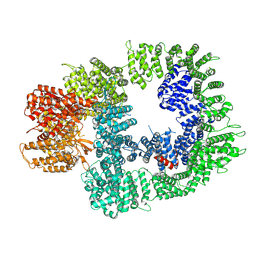 | | Cryo-EM structure of DNA-PKcs in complex with NU7441 | | Descriptor: | 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
5ZB2
 
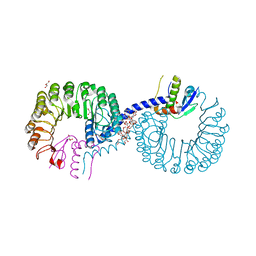 | | Crystal structure of Rad7 and Elc1 complex in yeast | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Jiang, T, Liu, L, Huo, Y. | | Deposit date: | 2018-02-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.30000544 Å) | | Cite: | Crystal structure of the yeast Rad7-Elc1 complex and assembly of the Rad7-Rad16-Elc1-Cul3 complex.
DNA Repair (Amst.), 77, 2019
|
|
7COC
 
 | | Ternary complex of DNA polymerase Mu (K438A/Q441A) with 1-nt gapped DNA (T:dGMPNPP) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*GP*GP*CP*TP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), ... | | Authors: | Guo, M, Zhao, Y. | | Deposit date: | 2020-08-04 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of genome instability mediated by human DNA polymerase mu misincorporation.
Nat Commun, 12, 2021
|
|
7COB
 
 | | Ternary complex of DNA polymerase Mu (Q441A) with 1-nt gapped DNA (T:dGMPNPP) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*GP*GP*CP*TP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), ... | | Authors: | Guo, M, Zhao, Y. | | Deposit date: | 2020-08-04 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Mechanism of genome instability mediated by human DNA polymerase mu misincorporation.
Nat Commun, 12, 2021
|
|
7CO6
 
 | |
7CO9
 
 | | Ternary complex of DNA polymerase Mu with 1-nt gapped DNA (T:dGMPNPP) and Mg | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*GP*GP*CP*TP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), ... | | Authors: | Guo, M, Zhao, Y. | | Deposit date: | 2020-08-04 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Mechanism of genome instability mediated by human DNA polymerase mu misincorporation.
Nat Commun, 12, 2021
|
|
7CO8
 
 | | Ternary complex of DNA polymerase Mu with 2-nt gapped DNA (T:dGMPNPP) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*GP*GP*CP*TP*TP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), ... | | Authors: | Guo, M, Zhao, Y. | | Deposit date: | 2020-08-04 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Mechanism of genome instability mediated by human DNA polymerase mu misincorporation.
Nat Commun, 12, 2021
|
|
7COD
 
 | |
7COA
 
 | | Ternary complex of DNA polymerase Mu with 1-nt gapped DNA (T:dGMPNPP) and Mn | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*GP*GP*CP*TP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), ... | | Authors: | Guo, M, Zhao, Y. | | Deposit date: | 2020-08-04 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Mechanism of genome instability mediated by human DNA polymerase mu misincorporation.
Nat Commun, 12, 2021
|
|
6DBL
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBQ
 
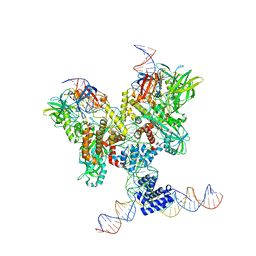 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
