2WZK
 
 | | Structure of the Cul5 N-terminal domain at 2.05A resolution. | | Descriptor: | 1,2-ETHANEDIOL, CULLIN-5 | | Authors: | Muniz, J.R.C, Ayinampudi, V, Zhang, Y, Babon, J.J, Chaikuad, A, Krojer, T, Pike, A.C.W, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bullock, A.N. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Architecture of the Ankyrin Socs Box Family of Cul5-Dependent E3 Ubiquitin Ligases
J.Mol.Biol., 425, 2013
|
|
8EI2
 
 | | Crystal structure of the N-terminal domain of CUL5 in complex with H314, a Helicon Polypeptide | | Descriptor: | Cullin-5, H314, N,N'-(1,4-phenylene)diacetamide | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8EI1
 
 | | Crystal structure of the N-terminal domain of CUL4B in complex with H316, a Helicon Polypeptide | | Descriptor: | Cullin-4B, H316, N,N'-(1,4-phenylene)diacetamide | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
4A64
 
 | | Crystal structure of the N-terminal domain of human Cul4B at 2.57A resolution | | Descriptor: | 1,2-ETHANEDIOL, CULLIN-4B | | Authors: | Vollmar, M, Ayinampudi, V, Cooper, C, Guo, K, Krojer, T, Muniz, J.R.C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A, Bullock, A. | | Deposit date: | 2011-10-31 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure of the N-Terminal Domain of Human Cul4B at 2.57A Resolution
To be Published
|
|
1LDJ
 
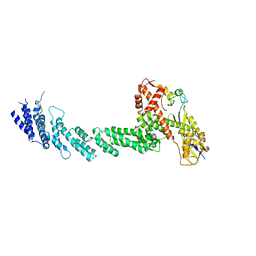 | | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF Ubiquitin Ligase Complex | | Descriptor: | Cullin homolog 1, ZINC ION, ring-box protein 1 | | Authors: | Zheng, N, Schulman, B.A, Song, L, Miller, J.J, Jeffrey, P.D, Wang, P, Chu, C, Koepp, D.M, Elledge, S.J, Pagano, M, Conaway, R.C, Conaway, J.W, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex.
Nature, 416, 2002
|
|
1U6G
 
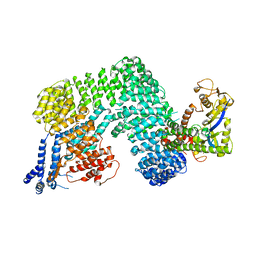 | | Crystal Structure of The Cand1-Cul1-Roc1 Complex | | Descriptor: | Cullin homolog 1, RING-box protein 1, TIP120 protein, ... | | Authors: | Goldenberg, S.J, Shumway, S.D, Cascio, T.C, Garbutt, K.C, Liu, J, Xiong, Y, Zheng, N. | | Deposit date: | 2004-07-29 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases
Cell(Cambridge,Mass.), 119, 2004
|
|
3RTR
 
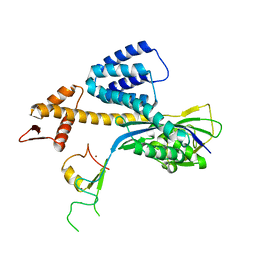 | | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Calabrese, M.F, Scott, D.C, Duda, D.M, Grace, C.R, Kurinov, I, Kriwacki, R.W, Schulman, B.A. | | Deposit date: | 2011-05-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2HYE
 
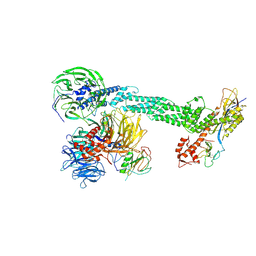 | | Crystal Structure of the DDB1-Cul4A-Rbx1-SV5V Complex | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, Nonstructural protein V, ... | | Authors: | Angers, S, Li, T, Yi, X, MacCoss, M.J, Moon, R.T, Zheng, N. | | Deposit date: | 2006-08-05 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery.
Nature, 443, 2006
|
|
1LDK
 
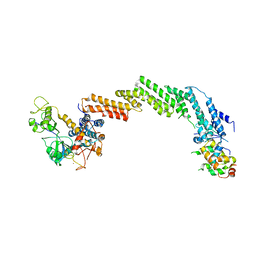 | | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF Ubiquitin Ligase Complex | | Descriptor: | CULLIN HOMOLOG, CYCLIN A/CDK2-ASSOCIATED PROTEIN P19, SKP2-like protein type gamma, ... | | Authors: | Zheng, N, Schulman, B.A, Song, L, Miller, J.J, Jeffrey, P.D, Wang, P, Chu, C, Koepp, D.M, Elledge, S.J, Pagano, M, Conaway, R.C, Conaway, J.W, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex.
Nature, 416, 2002
|
|
6V9I
 
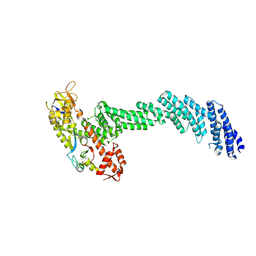 | | cryo-EM structure of Cullin5 bound to RING-box protein 2 (Cul5-Rbx2) | | Descriptor: | Immunoglobulin G-binding protein G,Cullin-5, RING-box protein 2, ZINC ION | | Authors: | Komives, E.A, Lumpkin, R.J, Baker, R.W, Leschziner, A.E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structure and dynamics of the ASB9 CUL-RING E3 Ligase.
Nat Commun, 11, 2020
|
|
3DPL
 
 | |
4F52
 
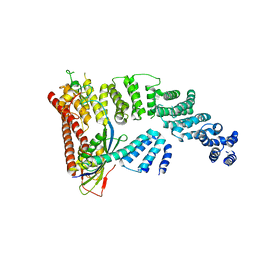 | | Structure of a Glomulin-RBX1-CUL1 complex | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, Glomulin, ... | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2012-05-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a Glomulin-RBX1-CUL1 Complex: Inhibition of a RING E3 Ligase through Masking of Its E2-Binding Surface.
Mol.Cell, 47, 2012
|
|
7OKQ
 
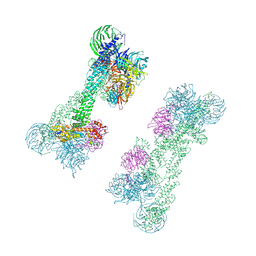 | | Cryo-EM Structure of the DDB1-DCAF1-CUL4A-RBX1 Complex | | Descriptor: | Cullin-4A, DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, ... | | Authors: | Mohamed, W.I, Schenk, A.D, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The CRL4 DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state.
Embo J., 40, 2021
|
|
8CDJ
 
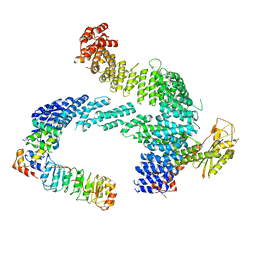 | | CAND1 b-hairpin++-SCF-SKP2 CAND1 rolling SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2023-01-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
8CDK
 
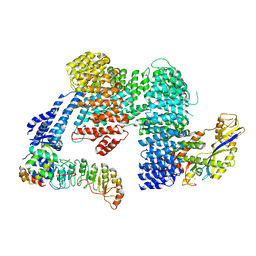 | |
4A0C
 
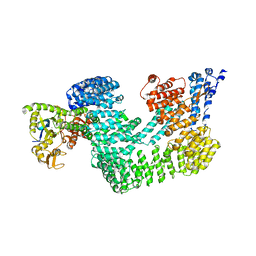 | | Structure of the CAND1-CUL4B-RBX1 complex | | Descriptor: | CULLIN-4B, CULLIN-ASSOCIATED NEDD8-DISSOCIATED PROTEIN 1, E3 UBIQUITIN-PROTEIN LIGASE RBX1, ... | | Authors: | Scrima, A, Fischer, E.S, Faty, M, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
8RHZ
 
 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2023-12-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7E
 
 | |
8VVY
 
 | | Human Cullin-1 in complex with CAND2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 2 | | Authors: | Kenny, S, Liu, X, Das, C. | | Deposit date: | 2024-01-31 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Cryo-EM structure of Human CAND2 in complex with Cullin-1
To Be Published
|
|
7Z8R
 
 | | CAND1-CUL1-RBX1 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7ZBZ
 
 | |
7Z8V
 
 | | CAND1-SCF-SKP2 (SKP1deldel) CAND1 engaged SCF rocked | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7Z8T
 
 | | CAND1-SCF-SKP2 CAND1 engaged SCF rocked | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7ZBW
 
 | | CAND1-SCF-SKP2 CAND1 rolling-2 SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
8U80
 
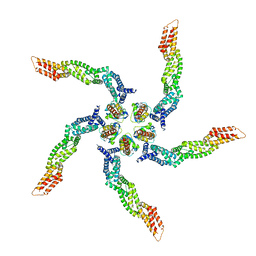 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(BTB)/Cullin3(NTD) | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3 | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
