1TO5
 
 | | Structure of the cytosolic Cu,Zn SOD from S. mansoni | | Descriptor: | ACETATE ION, COPPER (II) ION, Superoxide dismutase, ... | | Authors: | Cardoso, R.M.F, Silva, C.H.T.P, Ulian de Araujo, A.P, Tanaka, T, Tanaka, M, Garratt, R.C. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the cytosolic Cu,Zn superoxide dismutase from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TO6
 
 | | Glycerate kinase from Neisseria meningitidis (serogroup A) | | Descriptor: | Glycerate kinase, SULFATE ION | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-13 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glycerate kinase from Neisseria meningitidis (serogroup A)
To be Published
|
|
1TO9
 
 | |
1TOA
 
 | | PERIPLASMIC ZINC BINDING PROTEIN TROA FROM TREPONEMA PALLIDUM | | Descriptor: | GLYCEROL, PROTEIN (PERIPLASMIC BINDING PROTEIN TROA), ZINC ION | | Authors: | Lee, Y.H, Deka, R.K, Norgard, M.V, Radolf, J.D, Hasemann, C.A. | | Deposit date: | 1999-03-03 | | Release date: | 1999-07-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone.
Nat.Struct.Biol., 6, 1999
|
|
1TOB
 
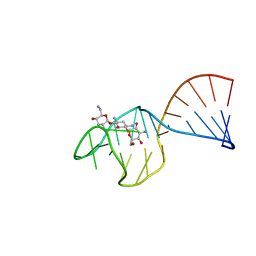 | | SACCHARIDE-RNA RECOGNITION IN AN AMINOGLYCOSIDE ANTIBIOTIC-RNA APTAMER COMPLEX, NMR, 7 STRUCTURES | | Descriptor: | 1,3-DIAMINO-5,6-DIHYDROXYCYCLOHEXANE, 2,6-diammonio-2,3,6-trideoxy-alpha-D-glucopyranose, 3-ammonio-3-deoxy-alpha-D-glucopyranose, ... | | Authors: | Jiang, L, Suri, A.K, Fiala, R, Patel, D.J. | | Deposit date: | 1996-12-12 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in an aminoglycoside antibiotic-RNA aptamer complex.
Chem.Biol., 4, 1997
|
|
1TOC
 
 | |
1TOE
 
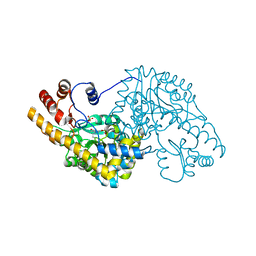 | | Unliganded structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, SULFATE ION | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOF
 
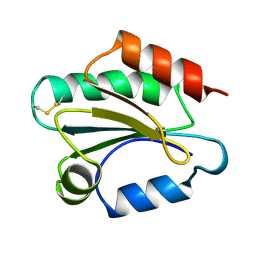 | | THIOREDOXIN H (OXIDIZED FORM), NMR, 23 STRUCTURES | | Descriptor: | THIOREDOXIN H | | Authors: | Mittard, V, Blackledge, M.J, Stein, M, Jacquot, J.-P, Marion, D, Lancelin, J.-M. | | Deposit date: | 1996-05-30 | | Release date: | 1996-12-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxidised thioredoxin h from the eukaryotic green alga Chlamydomonas reinhardtii.
Eur.J.Biochem., 243, 1997
|
|
1TOG
 
 | | Hydrocinnamic acid-bound structure of SRHEPT + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOH
 
 | |
1TOI
 
 | | Hydrocinnamic acid-bound structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOJ
 
 | | Hydrocinnamic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOK
 
 | | Maleic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, MALEIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOL
 
 | | FUSION OF N-TERMINAL DOMAIN OF THE MINOR COAT PROTEIN FROM GENE III IN PHAGE M13, AND C-TERMINAL DOMAIN OF E. COLI PROTEIN-TOLA | | Descriptor: | PROTEIN (FUSION PROTEIN CONSISTING OF MINOR COAT PROTEIN, GLYCINE RICH LINKER, TOLA, ... | | Authors: | Lubkowski, J, Wlodawer, A, Hennecke, F, Plueckthun, A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA.
Structure Fold.Des., 7, 1999
|
|
1TOM
 
 | | ALPHA-THROMBIN COMPLEXED WITH HIRUGEN | | Descriptor: | ALPHA-THROMBIN, HIRUGEN, METHYL-PHE-PRO-AMINO-CYCLOHEXYLGLYCINE | | Authors: | Chen, Z. | | Deposit date: | 1996-12-06 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, evaluation, and crystallographic analysis of L-371,912: A potent and selective active-site thrombin inhibitor.
Bioorg.Med.Chem.Lett., 7, 1997
|
|
1TON
 
 | |
1TOO
 
 | | Interleukin 1B Mutant F146W | | Descriptor: | Interleukin-1 beta | | Authors: | Adamek, D.H, Guerrero, L, Caspar, D.L. | | Deposit date: | 2004-06-14 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and energetic consequences of mutations in a solvated hydrophobic cavity.
J.Mol.Biol., 346, 2005
|
|
1TOP
 
 | |
1TOQ
 
 | | CRYSTAL STRUCTURE OF A GALACTOSE SPECIFIC LECTIN FROM ARTOCARPUS HIRSUTA IN COMPLEX WITH METHYL-a-D-GALACTOSE | | Descriptor: | AGGLUTININ ALPHA CHAIN, AGGLUTININ BETA CHAIN, methyl alpha-D-galactopyranoside | | Authors: | Rao, K.N, Suresh, C.G, Katre, U.V, Gaikwad, S.M, Khan, M.I. | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two orthorhombic crystal structures of a galactose-specific lectin from Artocarpus hirsuta in complex with methyl-alpha-D-galactose.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TOR
 
 | | MOLECULAR DYNAMICS SIMULATION FROM 2D-NMR DATA OF THE FREE ACHR MIR DECAPEPTIDE AND THE ANTIBODY-BOUND [A76]MIR ANALOGUE | | Descriptor: | ACETYLCHOLINE RECEPTOR, MAIN IMMUNOGENIC REGION | | Authors: | Orlewski, P, Tsikaris, V, Sakarellos, C, Sakarellos-Daistiotis, M, Vatzaki, E, Tzartos, S.J, Marraud, M, Cung, M.T. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Compared structures of the free nicotinic acetylcholine receptor main immunogenic region (MIR) decapeptide and the antibody-bound [A76]MIR analogue: a molecular dynamics simulation from two-dimensional NMR data.
Biopolymers, 40, 1996
|
|
1TOS
 
 | | TORPEDO CALIFORNICA ACHR RECEPTOR [ALA76] ANALOGUE COMPLEXED WITH THE ANTI-ACETYLCHOLINE MAB6 MONOCLONAL ANTIBODY | | Descriptor: | ACETYLCHOLINE RECEPTOR [ALA76] MIR ANALOGUE | | Authors: | Orlewski, P, Tsikaris, V, Sakarellos, C, Sakarellos-Daistiotis, M, Vatzaki, E, Tzartos, S.J, Marraud, M, Cung, M.T. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Compared structures of the free nicotinic acetylcholine receptor main immunogenic region (MIR) decapeptide and the antibody-bound [A76]MIR analogue: a molecular dynamics simulation from two-dimensional NMR data.
Biopolymers, 40, 1996
|
|
1TOT
 
 | | ZZ Domain of CBP- a Novel Fold for a Protein Interaction Module | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Legge, G.B, Martinez-Yamout, M.A, Hambly, D.M, Trinh, T, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-06-15 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module
J.Mol.Biol., 343, 2004
|
|
1TOU
 
 | | Crystal structure of human adipocyte fatty acid binding protein in complex with a non-covalent ligand | | Descriptor: | 2-[(2-OXO-2-PIPERIDIN-1-YLETHYL)SULFANYL]-6-(TRIFLUOROMETHYL)PYRIMIDIN-4-OL, Fatty acid-binding protein, adipocyte | | Authors: | Ringom, R, Axen, E, Uppenberg, J, Lundback, T, Rondahl, L, Barf, T. | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substituted benzylamino-6-(trifluoromethyl)pyrimidin-4(1H)-ones: a novel class of selective human A-FABP inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1TOV
 
 | | Structural genomics of Caenorhabditis elegans: CAP-GLY domain of F53F4.3 | | Descriptor: | Hypothetical protein F53F4.3 in chromosome V, SULFATE ION | | Authors: | Li, S, Finley, J, Liu, Z.J, Qiu, S.H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, Delucas, L.J, Richardson, D, Richardson, J, Wang, B.C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-06-15 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the Cytoskeleton-Associated Protein Glycine-Rich (CAP-Gly) Domain
J.Biol.Chem., 277, 2002
|
|
1TOW
 
 | | Crystal structure of human adipocyte fatty acid binding protein in complex with a carboxylic acid ligand | | Descriptor: | 4-(9H-CARBAZOL-9-YL)BUTANOIC ACID, Fatty acid-binding protein, adipocyte | | Authors: | Lehmann, F, Haile, S, Axen, E, Medina, C, Uppenberg, J, Svensson, S, Lundback, T, Rondahl, L, Barf, T. | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of inhibitors of human adipocyte fatty acid-binding protein, a potential type 2 diabetes target.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
