1LKF
 
 | | LEUKOCIDIN F (HLGB) FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | LEUKOCIDIN F SUBUNIT | | Authors: | Olson, R, Nariya, H, Yokota, K, Kamio, Y, Gouaux, J.E. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel.
Nat.Struct.Biol., 6, 1999
|
|
1LKI
 
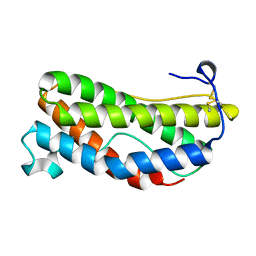 | | THE CRYSTAL STRUCTURE AND BIOLOGICAL FUNCTION OF LEUKEMIA INHIBITORY FACTOR: IMPLICATIONS FOR RECEPTOR BINDING | | Descriptor: | LEUKEMIA INHIBITORY FACTOR | | Authors: | Robinson, R.C, Grey, L.M, Staunton, D, Stuart, D.I, Heath, J.K, Jones, E.Y. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure and biological function of leukemia inhibitory factor: implications for receptor binding.
Cell(Cambridge,Mass.), 77, 1994
|
|
1LKJ
 
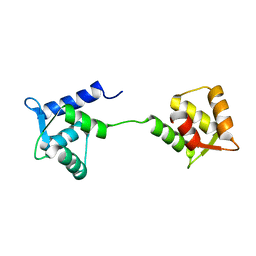 | | NMR Structure of Apo Calmodulin from Yeast Saccharomyces cerevisiae | | Descriptor: | Calmodulin | | Authors: | Ishida, H, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2002-04-25 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of apocalmodulin from Saccharomyces cerevisiae implies a mechanism for its unique Ca2+ binding property.
Biochemistry, 41, 2002
|
|
1LKK
 
 | |
1LKL
 
 | |
1LKM
 
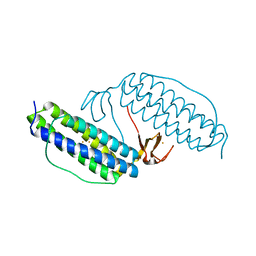 | | Crystal structure of Desulfovibrio vulgaris rubrerythrin all-iron(III) form | | Descriptor: | FE (III) ION, Rubrerythrin all-iron(III) form | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.J, Rose, J, Wang, B.C. | | Deposit date: | 2002-04-25 | | Release date: | 2002-09-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | X-ray Crystal Structures of Reduced Rubrerythrin and its Azide Adduct: A Structure-Based Mechanism for a Non-Heme DiIron Peroxidase
J.Am.Chem.Soc., 124, 2002
|
|
1LKN
 
 | |
1LKO
 
 | | Crystal structure of Desulfovibrio vulgaris rubrerythrin all-iron(II) form | | Descriptor: | FE (II) ION, Rubrerythrin all-iron(II) form | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.J, Rose, J, Wang, B.C. | | Deposit date: | 2002-04-25 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray Crystal Structures of Reduced Rubrerythrin and its Azide Adduct: A Structure-Based Mechanism for a Non-Heme DiIron Peroxidase
J.Am.Chem.Soc., 124, 2002
|
|
1LKP
 
 | | Crystal structure of Desulfovibrio vulgaris rubrerythrin all-iron(II) form, azide adduct | | Descriptor: | AZIDE ION, FE (II) ION, Rubrerythrin | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.J, Rose, J, Wang, B.C. | | Deposit date: | 2002-04-25 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | X-ray Crystal Structures of Reduced Rubrerythrin and its Azide Adduct: A Structure-Based Mechanism for a Non-Heme Diiron Peroxidase
J.Am.Chem.Soc., 124, 2002
|
|
1LKQ
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-GLY, VAL-A3-GLY, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 20 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Weiss, M.A, Hua, Q.X, Chu, Y.C, Jia, W, Philips, N.F, Wang, R.Y, Katsoyannis, P.G. | | Deposit date: | 2002-04-25 | | Release date: | 2002-05-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Mechanism of insulin chain combination. Asymmetric roles of A-chain alpha-helices in disulfide pairing
J.Biol.Chem., 277, 2002
|
|
1LKR
 
 | | MONOCLINIC HEN EGG WHITE LYSOZYME IODIDE | | Descriptor: | IODIDE ION, LYSOZYME | | Authors: | Steinrauf, L.K. | | Deposit date: | 1998-01-20 | | Release date: | 1998-04-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of monoclinic lysozyme iodide at 1.6 A and of triclinic lysozyme nitrate at 1.1 A.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1LKS
 
 | | HEN EGG WHITE LYSOZYME NITRATE | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Steinrauf, L.K. | | Deposit date: | 1997-10-09 | | Release date: | 1998-04-29 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures of monoclinic lysozyme iodide at 1.6 A and of triclinic lysozyme nitrate at 1.1 A.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1LKT
 
 | | CRYSTAL STRUCTURE OF THE HEAD-BINDING DOMAIN OF PHAGE P22 TAILSPIKE PROTEIN | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S. | | Deposit date: | 1997-10-17 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage.
J.Mol.Biol., 267, 1997
|
|
1LKV
 
 | |
1LKX
 
 | | MOTOR DOMAIN OF MYOE, A CLASS-I MYOSIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN IE HEAVY CHAIN, ... | | Authors: | Kollmar, M, Durrwang, U, Kliche, W, Manstein, D.J, Kull, F.J. | | Deposit date: | 2002-04-26 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the motor domain of a class-I myosin.
EMBO J., 21, 2002
|
|
1LKY
 
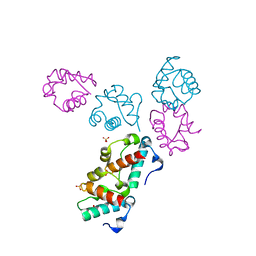 | | Structure of the wild-type TEL-SAM polymer | | Descriptor: | SULFATE ION, TRANSCRIPTION FACTOR ETV6 | | Authors: | Tran, H.H, Kim, C.A, Faham, S, Bowie, J.U. | | Deposit date: | 2002-04-26 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Native interface of the SAM domain polymer of TEL.
BMC STRUCT.BIOL., 2, 2002
|
|
1LKZ
 
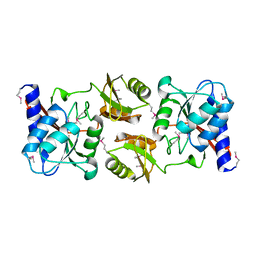 | | Crystal structure of D-ribose-5-phosphate isomerase (RpiA) from Escherichia coli. | | Descriptor: | Ribose 5-phosphate isomerase A | | Authors: | Rangarajan, E.S, Sivaraman, J, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-04-26 | | Release date: | 2002-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of D-ribose-5-phosphate isomerase (RpiA) from Escherichia coli
Proteins, 48, 2002
|
|
1LL0
 
 | |
1LL1
 
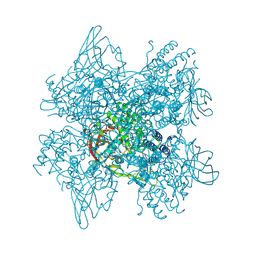 | | HYDROXO BRIDGE MET FORM HEMOCYANIN FROM LIMULUS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, METCYANIN II | | Authors: | Liu, S, Magnus, K. | | Deposit date: | 1997-03-17 | | Release date: | 1997-08-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic Studies of Hydroxo, Nitrosyl, Azido and Fluro met Form Hemocyanin Subunit II from Limulus
To be Published
|
|
1LL2
 
 | | Crystal Structure of Rabbit Muscle Glycogenin Complexed with UDP-glucose and Manganese | | Descriptor: | GLYCOGENIN-1, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Gibbons, B.J, Roach, P.J, Hurley, T.D. | | Deposit date: | 2002-04-26 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the autocatalytic initiator of glycogen biosynthesis, glycogenin.
J.Mol.Biol., 319, 2002
|
|
1LL3
 
 | |
1LL4
 
 | | STRUCTURE OF C. IMMITIS CHITINASE 1 COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE 1 | | Authors: | Bortone, K, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2002-04-26 | | Release date: | 2002-09-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | THE STRUCTURE OF AN ALLOSAMIDIN COMPLEX WITH THE Coccidioides IMMITIS CHITINASE DEFINES A ROLE FOR A SECOND ACID RESIDUE IN SUBSTRATE-ASSISTED MECHANISM
J.Mol.Biol., 320, 2002
|
|
1LL5
 
 | |
1LL6
 
 | |
1LL7
 
 | |
