1C3B
 
 | | AMPC BETA-LACTAMASE FROM E. COLI COMPLEXED WITH INHIBITOR, BENZO(B)THIOPHENE-2-BORONIC ACID (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, CEPHALOSPORINASE | | Authors: | Powers, R.A, Blazquez, J, Weston, G.S, Morosini, M.I, Baquero, F, Shoichet, B.K. | | Deposit date: | 1999-07-27 | | Release date: | 1999-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The complexed structure and antimicrobial activity of a non-beta-lactam inhibitor of AmpC beta-lactamase.
Protein Sci., 8, 1999
|
|
1BOB
 
 | | HISTONE ACETYLTRANSFERASE HAT1 FROM SACCHAROMYCES CEREVISIAE IN COMPLEX WITH ACETYL COENZYME A | | Descriptor: | ACETYL COENZYME *A, CALCIUM ION, HISTONE ACETYLTRANSFERASE | | Authors: | Dutnall, R.N, Tafrov, S.T, Sternglanz, R, Ramakrishnan, V. | | Deposit date: | 1998-07-02 | | Release date: | 1999-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily.
Cell(Cambridge,Mass.), 94, 1998
|
|
1C3J
 
 | | T4 PHAGE BETA-GLUCOSYLTRANSFERASE: SUBSTRATE BINDING AND PROPOSED CATALYTIC MECHANISM | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Imberty, A, Aschke-Sonnenborn, U, Ruger, W, Freemont, P.S. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | T4 phage beta-glucosyltransferase: substrate binding and proposed catalytic mechanism.
J.Mol.Biol., 292, 1999
|
|
1C3Q
 
 | | CRYSTAL STRUCTURE OF NATIVE THIAZOLE KINASE IN THE MONOCLINIC FORM | | Descriptor: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, CHLORIDE ION, Hydroxyethylthiazole kinase | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
1BKE
 
 | | HUMAN SERUM ALBUMIN IN A COMPLEX WITH MYRISTIC ACID AND TRI-IODOBENZOIC ACID | | Descriptor: | 2,3,5-TRIIODOBENZOIC ACID, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Curry, S, Mandelkow, H, Brick, P, Franks, N. | | Deposit date: | 1998-07-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites.
Nat.Struct.Biol., 5, 1998
|
|
1BOL
 
 | |
1BTW
 
 | |
1BVL
 
 | | HUMANIZED ANTI-LYSOZYME FV | | Descriptor: | HULYS11 | | Authors: | Holmes, M.A, Foote, J. | | Deposit date: | 1998-09-16 | | Release date: | 1999-02-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural consequences of humanizing an antibody.
J.Immunol., 158, 1997
|
|
1BP7
 
 | | GROUP I MOBILE INTRON ENDONUCLEASE I-CREI COMPLEXED WITH HOMING SITE DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP* GP*C)-3'), DNA (5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP* CP*G)-3'), ... | | Authors: | Jurica, M.S, Monnat Junior, R.J, Stoddard, B.L. | | Deposit date: | 1998-08-13 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI.
Mol.Cell, 2, 1998
|
|
1BQ1
 
 | | E. COLI THYMIDYLATE SYNTHASE MUTANT N177A IN COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Reyes, C.L, Sage, C.R, Rutenber, E.E, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-20 | | Release date: | 1999-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inactivity of N229A thymidylate synthase due to water-mediated effects: isolating a late stage in methyl transfer.
J.Mol.Biol., 284, 1998
|
|
1BM2
 
 | |
1BM9
 
 | |
1BRQ
 
 | |
1BN6
 
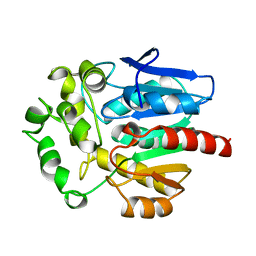 | | HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1998-07-31 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
1BS7
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM | | Descriptor: | NICKEL (II) ION, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
1BSI
 
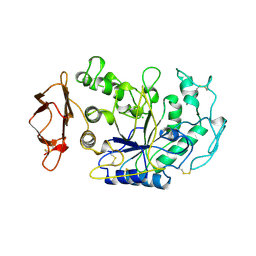 | | HUMAN PANCREATIC ALPHA-AMYLASE FROM PICHIA PASTORIS, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Rydberg, E.H, Sidhu, G, Vo, H.C, Hewitt, J, Cote, H.C.F, Wang, Y, Numao, S, Macgillivray, R.T.A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 1998-08-28 | | Release date: | 1999-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cloning, mutagenesis, and structural analysis of human pancreatic alpha-amylase expressed in Pichia pastoris.
Protein Sci., 8, 1999
|
|
1BT2
 
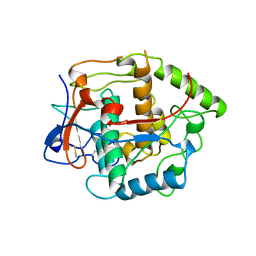 | | CATECHOL OXIDASE FROM IPOMOEA BATATAS (SWEET POTATOES) IN THE REDUCED CU(I)-CU(I) STATE | | Descriptor: | CU-O-CU LINKAGE, PROTEIN (CATECHOL OXIDASE) | | Authors: | Klabunde, T, Eicken, C, Sacchettini, J.C, Krebs, B. | | Deposit date: | 1998-09-02 | | Release date: | 1999-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a plant catechol oxidase containing a dicopper center.
Nat.Struct.Biol., 5, 1998
|
|
1BRE
 
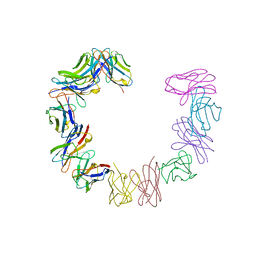 | | IMMUNOGLOBULIN LIGHT CHAIN PROTEIN | | Descriptor: | BENCE-JONES KAPPA I PROTEIN BRE | | Authors: | Schormann, N, Benson, M.D. | | Deposit date: | 1995-07-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tertiary structure of an amyloid immunoglobulin light chain protein: a proposed model for amyloid fibril formation.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1BQ4
 
 | |
1BRG
 
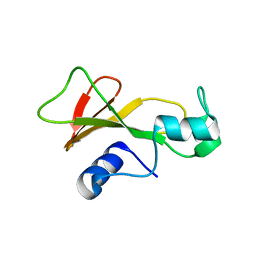 | |
1BRU
 
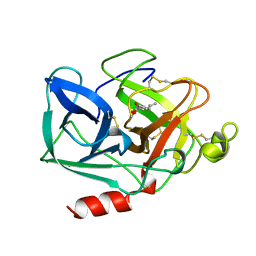 | |
1BUH
 
 | |
1BTL
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI TEM1 BETA-LACTAMASE AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | BETA-LACTAMASE TEM1, SULFATE ION | | Authors: | Jelsch, C, Mourey, L, Masson, J.M, Samama, J.P. | | Deposit date: | 1993-11-01 | | Release date: | 1995-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Escherichia coli TEM1 beta-lactamase at 1.8 A resolution.
Proteins, 16, 1993
|
|
1BTZ
 
 | |
1BUP
 
 | | T13S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sousa, M.C, Mckay, D.B. | | Deposit date: | 1998-09-03 | | Release date: | 1998-09-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change.
Biochemistry, 37, 1998
|
|
