8DY6
 
 | | Vaccine elicited Antibody MU89+S27Y bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-08-03 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
6M6C
 
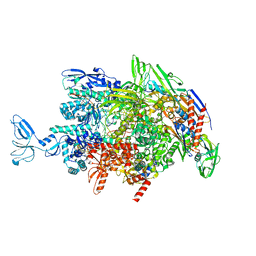 | | CryoEM structure of Thermus thermophilus RNA polymerase elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
7M7F
 
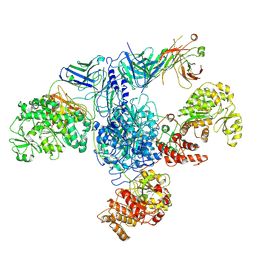 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 1 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
6UG1
 
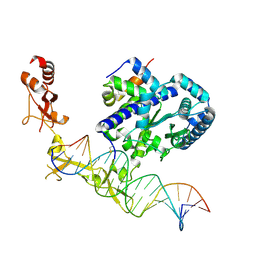 | | Sequence impact in DNA duplex opening by the Rad4/XPC nucleotide excision repair complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Paul, D, Min, J.-H. | | Deposit date: | 2019-09-25 | | Release date: | 2021-03-31 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | Impact of DNA sequences on DNA 'opening' by the Rad4/XPC nucleotide excision repair complex.
DNA Repair (Amst), 107, 2021
|
|
6PIV
 
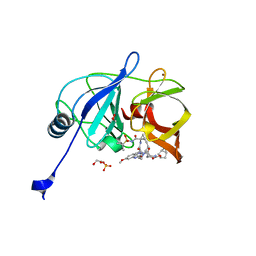 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-7 (NR03-77) | | Descriptor: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
8E0O
 
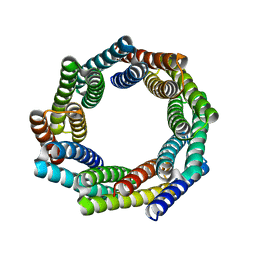 | |
7M7H
 
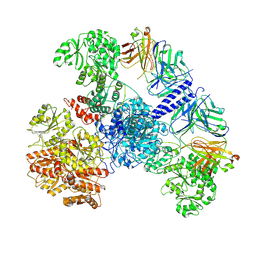 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 1' | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
5PY4
 
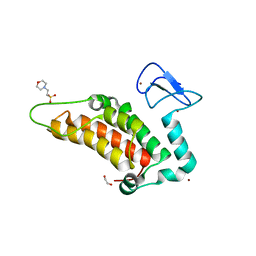 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 64) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6ZMQ
 
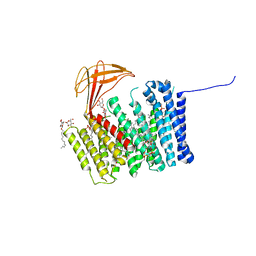 | | Cytochrome c Heme Lyase CcmF | | Descriptor: | Cytochrome C-type biogenesis protein ccmF, DODECYL-BETA-D-MALTOSIDE, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Brausemann, A, Einsle, O. | | Deposit date: | 2020-07-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Architecture of the membrane-bound cytochrome c heme lyase CcmF.
Nat.Chem.Biol., 17, 2021
|
|
6PJ0
 
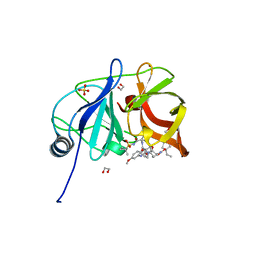 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-5 (NR01-97) | | Descriptor: | 1,2-ETHANEDIOL, 1-ethylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4 Aprotease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6UMA
 
 | |
8EBT
 
 | |
6M7X
 
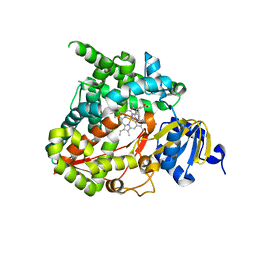 | | Structure of human CYP11B1 in complex with fadrozole | | Descriptor: | 4-[(5S)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-5-yl]benzonitrile, Cytochrome P450 11B1, mitochondrial, ... | | Authors: | Scott, E.E, Brixius-Anderko, S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Structure of human cortisol-producing cytochrome P450 11B1 bound to the breast cancer drug fadrozole provides insights for drug design.
J. Biol. Chem., 294, 2019
|
|
6ZOC
 
 | | Erythromycin binding to the access pocket of AcrB-G616P L protomer and 3-formylrifamycin SV binding to the access pocket of AcrB-G616P T protomer | | Descriptor: | (2S,12Z,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-8-formyl-5,6,9,17,19-pentahydroxy-23-methoxy-2,4,12,16,18,20,22-heptam ethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-21-yl acetate, 1,2-ETHANEDIOL, DARPIN, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
8DNH
 
 | | Cryo-EM structure of nonmuscle beta-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
7ML5
 
 | | Structure of the Starch Branching Enzyme I (BEI) complexed with maltododecaose from Oryza sativa L | | Descriptor: | Isoform 2 of 1,4-alpha-glucan-branching enzyme, chloroplastic/amyloplastic, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Nayebi Gavgani, H, Fawaz, R, Geiger, J.H. | | Deposit date: | 2021-04-27 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A structural explanation for the mechanism and specificity of plant branching enzymes I and IIb.
J.Biol.Chem., 298, 2021
|
|
6P8D
 
 | |
6UMB
 
 | |
6M80
 
 | | Collagen peptide containing aza-proline and aza-glycine | | Descriptor: | 1,2-ETHANEDIOL, Collagen peptide containing aza-proline and aza-glycine, SULFATE ION | | Authors: | Chenoweth, D.M, Kasznel, A.J, Porter, N.J. | | Deposit date: | 2018-08-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Aza-proline effectively mimics l-proline stereochemistry in triple helical collagen.
Chem Sci, 10, 2019
|
|
5PYM
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 82) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
7M7G
 
 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 2 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
5PZ0
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 96) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6UJ8
 
 | | Crystal structure of HLA-B*07:02 with wild-type IDH2 peptide | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, HLA class I histocompatibility antigen, ... | | Authors: | Miller, M.S, Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2019-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
6M86
 
 | |
6ZPN
 
 | | Crystal structure of Chaetomium thermophilum Raptor | | Descriptor: | WD_REPEATS_REGION domain-containing protein | | Authors: | Imseng, S, Boehm, R, Jakob, R.P, Hall, M.N, Hiller, S, Maier, T. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The dynamic mechanism of 4E-BP1 recognition and phosphorylation by mTORC1.
Mol.Cell, 81, 2021
|
|
