1UAT
 
 | | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J | | Descriptor: | Azurin iso-2, COPPER (II) ION, SULFATE ION | | Authors: | Inoue, T, Suzuki, S, Nisho, N, Yamaguchi, K, Kataoka, K, Tobari, J, Yong, X, Hamanaka, S, Matsumura, H, Kai, Y. | | Deposit date: | 2003-03-19 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J
J.Mol.Biol., 333, 2003
|
|
1UF4
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase | | Descriptor: | N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid
To be published
|
|
1TOM
 
 | | ALPHA-THROMBIN COMPLEXED WITH HIRUGEN | | Descriptor: | ALPHA-THROMBIN, HIRUGEN, METHYL-PHE-PRO-AMINO-CYCLOHEXYLGLYCINE | | Authors: | Chen, Z. | | Deposit date: | 1996-12-06 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, evaluation, and crystallographic analysis of L-371,912: A potent and selective active-site thrombin inhibitor.
Bioorg.Med.Chem.Lett., 7, 1997
|
|
1UCW
 
 | |
1U8K
 
 | |
1U95
 
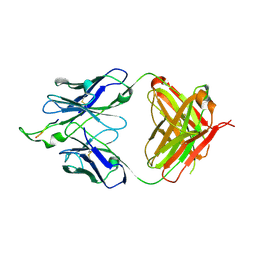 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDHWAS | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
1UGD
 
 | |
1TUD
 
 | |
1TZ3
 
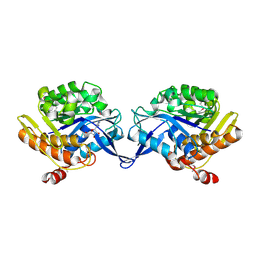 | | crystal structure of aminoimidazole riboside kinase complexed with aminoimidazole riboside | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOSIDE, POTASSIUM ION, putative sugar kinase | | Authors: | Zhang, Y, Dougherty, M, Downs, D.M, Ealick, S.E. | | Deposit date: | 2004-07-09 | | Release date: | 2004-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of an Aminoimidazole Riboside Kinase from Salmonella enterica; Implications for the Evolution of the Ribokinase Superfamily
STRUCTURE, 12, 2004
|
|
1TVU
 
 | |
1UCF
 
 | | The Crystal Structure of DJ-1, a Protein Related to Male Fertility and Parkinson's Disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Honbou, K, Suzuki, N.N, Horiuchi, M, Niki, T, Taira, T, Ariga, H, Inagaki, F. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of DJ-1, a Protein Related to Male Fertility and Parkinson's Disease
J.BIOL.CHEM., 278, 2003
|
|
1U1M
 
 | |
1U1T
 
 | | Hfq protein from Pseudomonas aeruginosa. High-salt crystals | | Descriptor: | Hfq protein | | Authors: | Nikulin, A.D, Stolboushkina, E.A, Perederina, A.A, Vassilieva, I.M, Blaesi, U, Moll, I, Kachalova, G, Yokoyama, S, Vassylyev, D, Garber, M, Nikonov, S.V, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-16 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Pseudomonas aeruginosa Hfq protein.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1UET
 
 | |
1TZE
 
 | |
1U1R
 
 | |
1U2O
 
 | | Crystal Structure Of The N-Domain Of Grp94 Lacking The Charged Domain In Complex With Neca | | Descriptor: | Endoplasmin, N-ETHYL-5'-CARBOXAMIDO ADENOSINE, PENTAETHYLENE GLYCOL, ... | | Authors: | Soldano, K.L, Jivan, A, Nicchitta, C.V, Gewirth, D.T. | | Deposit date: | 2004-07-19 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the N-terminal domain of GRP94. Basis for ligand specificity and regulation
J.Biol.Chem., 278, 2003
|
|
1TTI
 
 | |
1R8I
 
 | | Crystal structure of TraC | | Descriptor: | TraC | | Authors: | Yeo, H.-J, Yuan, Q, Beck, M.R, Baron, C, Waksman, G. | | Deposit date: | 2003-10-24 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1R5J
 
 | | Crystal Structure of a Phosphotransacetylase from Streptococcus pyogenes | | Descriptor: | putative phosphotransacetylase | | Authors: | Xu, Q.S, Shin, D.H, Pufan, R, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-10-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a phosphotransacetylase from Streptococcus pyogenes.
Proteins, 55, 2004
|
|
1RA5
 
 | | Bacterial cytosine deaminase D314A mutant bound to 5-fluoro-4-(S)-hydroxyl-3,4-dihydropyrimidine. | | Descriptor: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION, ... | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
1RAI
 
 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1RBH
 
 | |
1RDO
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1REZ
 
 | | HUMAN LYSOZYME-N-ACETYLLACTOSAMINE COMPLEX | | Descriptor: | GLYCEROL, LYSOZYME, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Origin of carbohydrate recognition specificity of human lysozyme revealed by affinity labeling.
Biochemistry, 35, 1996
|
|
