1QBM
 
 | |
1QKU
 
 | | WILD TYPE ESTROGEN NUCLEAR RECEPTOR LIGAND BINDING DOMAIN COMPLEXED WITH ESTRADIOL | | Descriptor: | ESTRADIOL, ESTRADIOL RECEPTOR | | Authors: | Ruff, M, Gangloff, M, Eiler, S, Duclaud, S, Wurtz, J.M, Moras, D. | | Deposit date: | 1999-08-05 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a mutant hERalpha ligand-binding domain reveals key structural features for the mechanism of partial agonism.
J. Biol. Chem., 276, 2001
|
|
1QMF
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) ACYL-ENZYME COMPLEX | | Descriptor: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, CEFUROXIME (OCT-3-ENE FORM), PENICILLIN-BINDING PROTEIN 2X | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Penicillin Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance
J.Mol.Biol., 299, 2000
|
|
1QO7
 
 | | Structure of Aspergillus niger epoxide hydrolase | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Zou, J.-Y, Hallberg, B.M, Bergfors, T, Oesch, F, Arand, M, Mowbray, S.L, Jones, T.A. | | Deposit date: | 1999-11-04 | | Release date: | 2000-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Aspergillus Niger Epoxide Hydrolase at 1.8A Resolution: Implications for the Structure and Function of the Mammalian Microsomal Class of Epoxide Hydrolases
Structure, 8, 2000
|
|
1R5H
 
 | | Crystal Structure of MetAP2 complexed with A320282 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-(2S,3R)-3-AMINO-4-CYCLOHEXYL-2-HYDROXY-BUTANO-N-(4-METHYLPHENYL)HYDRAZIDE | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1R6X
 
 | | The Crystal Structure of a Truncated Form of Yeast ATP Sulfurylase, Lacking the C-Terminal APS Kinase-like Domain, in complex with Sulfate | | Descriptor: | ATP:sulfate adenylyltransferase, COBALT (II) ION, SULFATE ION | | Authors: | Lalor, D.J, Schnyder, T, Saridakis, V, Pilloff, D.E, Dong, A, Tang, H, Leyh, T.S, Pai, E.F. | | Deposit date: | 2003-10-17 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional analysis of a truncated form of Saccharomyces cerevisiae ATP sulfurylase: C-terminal domain essential for oligomer formation but not for activity
Protein Eng., 16, 2003
|
|
1SCR
 
 | | HIGH-RESOLUTION STRUCTURES OF SINGLE-METAL-SUBSTITUTED CONCANAVALIN A: THE CO,CA-PROTEIN AT 1.6 ANGSTROMS AND THE NI,CA-PROTEIN AT 2.0 ANGSTROMS | | Descriptor: | CALCIUM ION, CONCANAVALIN A, NICKEL (II) ION | | Authors: | Emmerich, C, Helliwell, J.R, Redshaw, M, Naismith, J.H, Harrop, S.J, Raftery, J, Kalb, A.J, Yariv, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 1993-12-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of single-metal-substituted concanavalin A: the Co,Ca-protein at 1.6 A and the Ni,Ca-protein at 2.0 A.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1S70
 
 | | Complex between protein ser/thr phosphatase-1 (delta) and the myosin phosphatase targeting subunit 1 (MYPT1) | | Descriptor: | 130 kDa myosin-binding subunit of smooth muscle myosin phophatase (M130), MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-beta (or delta) catalytic subunit, ... | | Authors: | Kerff, F, Terrak, M, Dominguez, R. | | Deposit date: | 2004-01-28 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of protein phosphatase 1 regulation
Nature, 429, 2004
|
|
1RZF
 
 | | Crystal structure of Human anti-HIV-1 GP120-reactive antibody E51 | | Descriptor: | Fab E51 heavy chain, Fab E51 light chain, GLYCEROL, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1S4P
 
 | | Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p: ternary complex with GDP/Mn and methyl-alpha-mannoside acceptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Lobsanov, Y.D, Romero, P.A, Sleno, B, Yu, B, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2004-01-16 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of Kre2p/Mnt1p: A YEAST {alpha}1,2-MANNOSYLTRANSFERASE INVOLVED IN MANNOPROTEIN BIOSYNTHESIS
J.Biol.Chem., 279, 2004
|
|
1RZZ
 
 | | PHOTOSYNTHETIC REACTION CENTER DOUBLE MUTANT FROM RHODOBACTER SPHAEROIDES WITH ASP L213 REPLACED WITH ASN AND ARG M233 REPLACED WITH CYS IN THE CHARGE-NEUTRAL DQAQB STATE (TETRAGONAL FORM) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Xu, Q, Axelrod, H.L, Abresch, E.C, Paddock, M.L, Okamura, M.Y, Feher, G. | | Deposit date: | 2003-12-29 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-Ray Structure Determination of Three Mutants of the Bacterial Photosynthetic Reaction Centers from Rb. sphaeroides; Altered Proton Transfer Pathways.
STRUCTURE, 12, 2004
|
|
1RXD
 
 | | Crystal structure of human protein tyrosine phosphatase 4A1 | | Descriptor: | protein tyrosine phosphatase type IVA, member 1; Protein tyrosine phosphatase IVA1 | | Authors: | Sun, J.P, Fedorov, A.A, Almo, S.C, Zhang, Z.Y, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-18 | | Release date: | 2004-12-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1S3M
 
 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, NICKEL (II) ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
1S5H
 
 | | Potassium Channel Kcsa-Fab Complex T75C mutant in K+ | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, DIACYL GLYCEROL, ... | | Authors: | Mackinnon, R, Zhou, M. | | Deposit date: | 2004-01-20 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A mutant KcsA K(+) channel with altered conduction properties and selectivity filter ion distribution.
J.Mol.Biol., 338, 2004
|
|
1SA5
 
 | | Rat protein farnesyltransferase complexed with FPP and BMS-214662 | | Descriptor: | 3-BENZYL-1-(1H-IMIDAZOL-4-YLMETHYL)-4-(THIEN-2-YLSULFONYL)-2,3,4,5-TETRAHYDRO-1H-1,4-BENZODIAZEPINE-7-CARBONITRILE, ACETIC ACID, FARNESYL DIPHOSPHATE, ... | | Authors: | Reid, T.S, Beese, L.S. | | Deposit date: | 2004-02-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Anticancer Clinical Candidates R115777 (Tipifarnib) and BMS-214662 Complexed with Protein Farnesyltransferase Suggest a Mechanism of FTI Selectivity.
Biochemistry, 43, 2004
|
|
1SAV
 
 | | HUMAN ANNEXIN V WITH PROLINE SUBSTITUTION BY THIOPROLINE | | Descriptor: | ANNEXIN V, CALCIUM ION | | Authors: | Medrano, F.J, Minks, C, Budisa, N, Huber, R. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal and molecular structure of human annexin V after refinement. Implications for structure, membrane binding and ion channel formation of the annexin family of proteins.
J.Mol.Biol., 223, 1992
|
|
1SCJ
 
 | |
1SEM
 
 | |
1SER
 
 | | THE 2.9 ANGSTROMS CRYSTAL STRUCTURE OF T. THERMOPHILUS SERYL-TRNA SYNTHETASE COMPLEXED WITH TRNA SER | | Descriptor: | PROTEIN (SERYL-TRNA SYNTHETASE (E.C.6.1.1.11)), TRNASER | | Authors: | Biou, S, Cusack, V, Yaremchuk, A, Tukalo, M. | | Deposit date: | 1994-02-21 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser).
Science, 263, 1994
|
|
1SIG
 
 | |
1SI2
 
 | |
1SEI
 
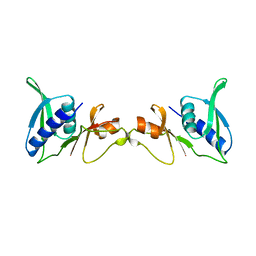 | | STRUCTURE OF 30S RIBOSOMAL PROTEIN S8 | | Descriptor: | RIBOSOMAL PROTEIN S8 | | Authors: | Davies, C, Ramakrishnan, V, White, S.W. | | Deposit date: | 1996-08-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evidence for specific S8-RNA and S8-protein interactions within the 30S ribosomal subunit: ribosomal protein S8 from Bacillus stearothermophilus at 1.9 A resolution.
Structure, 4, 1996
|
|
1RAH
 
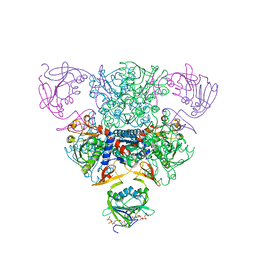 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1RDZ
 
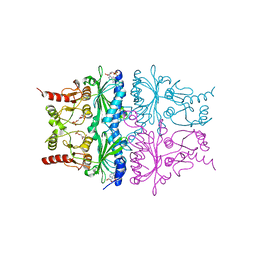 | | T-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
1RFS
 
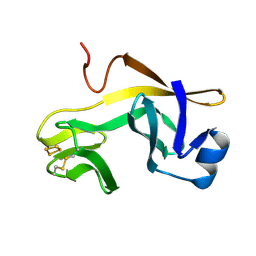 | | RIESKE SOLUBLE FRAGMENT FROM SPINACH | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, RIESKE PROTEIN | | Authors: | Carrell, C.J, Zhang, H, Cramer, W.A, Smith, J.L. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biological identity and diversity in photosynthesis and respiration: structure of the lumen-side domain of the chloroplast Rieske protein.
Structure, 5, 1997
|
|
