3MCS
 
 | |
3MZS
 
 | | Crystal Structure of Cytochrome P450 CYP11A1 in complex with 22-hydroxy-cholesterol | | Descriptor: | (3alpha,8alpha,22R)-cholest-5-ene-3,22-diol, Cholesterol side-chain cleavage enzyme, ISOPROPYL ALCOHOL, ... | | Authors: | Stout, C.D, Annalora, A, Mast, N, Pikuleva, I. | | Deposit date: | 2010-05-12 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Three-step Sequential Catalysis by the Cholesterol Side Chain Cleavage Enzyme CYP11A1.
J.Biol.Chem., 286, 2011
|
|
4H87
 
 | |
4HAQ
 
 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose and cellotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HD4
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium V218F mutant | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influencing the monophenolase/diphenolase activity ratio in tyrosinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4HD7
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium V218G mutant soaked in CuSO4 | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Influencing the monophenolase/diphenolase activity ratio in tyrosinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4GNF
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-15 | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GNG
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3K9me3 peptide | | Descriptor: | GLYCEROL, Histone H3.3, Histone-lysine N-methyltransferase NSD3, ... | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
3NFV
 
 | |
4H41
 
 | |
4H5J
 
 | |
3N91
 
 | |
4GX7
 
 | |
3N6Y
 
 | |
4HES
 
 | | Structure of a Beta-Lactamase Class A-like Protein from Veillonella parvula. | | Descriptor: | Beta-lactamase class A-like protein, FORMIC ACID, GLYCEROL, ... | | Authors: | Cuff, M.E, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-04 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Beta-Lactamase Class A-like Protein from Veillonella parvula.
TO BE PUBLISHED
|
|
4HFS
 
 | |
3NA6
 
 | |
4IAX
 
 | | Engineered human lipocalin 2 (CL31) in complex with Y-DTPA | | Descriptor: | N-{(1S,2S)-2-[bis(carboxymethyl)amino]cyclohexyl}-N-{(2R)-2-[bis(carboxymethyl)amino]-3-[4-({[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]carbamothioyl}amino)phenyl]propyl}glycine, Neutrophil gelatinase-associated lipocalin, YTTRIUM (III) ION | | Authors: | Eichinger, A, Skerra, A. | | Deposit date: | 2012-12-07 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided engineering of Anticalins with improved binding behavior and biochemical characteristics for application in radio-immuno imaging and/or therapy
J.Struct.Biol., 185, 2014
|
|
3NNR
 
 | |
3NPP
 
 | |
4HYZ
 
 | |
4IZT
 
 | |
4I8I
 
 | |
4I8T
 
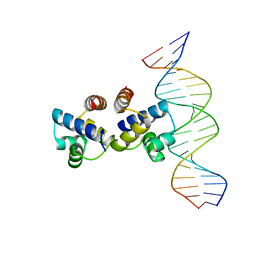 | | C.Esp1396I bound to a 19 base pair DNA duplex | | Descriptor: | DNA (5'-D(P*TP*GP*TP*GP*TP*GP*AP*TP*TP*AP*TP*AP*GP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(P*TP*GP*TP*TP*GP*AP*CP*TP*AP*TP*AP*AP*TP*CP*AP*CP*AP*CP*A)-3'), Regulatory protein | | Authors: | Martin, R.N.A, McGeehan, J.E, Kneale, G.G. | | Deposit date: | 2012-12-04 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of DNA-protein complexes regulating the restriction-modification system Esp1396I.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4IAS
 
 | |
