4HG7
 
 | | Crystal Structure of an MDM2/Nutlin-3a complex | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Noble, M.E.M, Anil, B, Riedinger, C, Endicott, J.A. | | Deposit date: | 2012-10-07 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of an MDM2-Nutlin-3a complex solved by the use of a validated MDM2 surface-entropy reduction mutant.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2ANQ
 
 | | Crystal Structure of E.coli DHFR in complex with NADPH and the inhibitor compound 10a. | | Descriptor: | (2,5-dimethylbenzene-1,4-diyl)dimethanediyl bis(N-carbamimidoylcarbamimidothioate), Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
1JNF
 
 | | Rabbit serum transferrin at 2.6 A resolution. | | Descriptor: | CARBONATE ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Hall, D.R, Hadden, J.M, Leonard, G.A, Bailey, S, Neu, M, Winn, M, Lindley, P.F. | | Deposit date: | 2001-07-24 | | Release date: | 2001-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal and molecular structures of diferric porcine and rabbit serum transferrins at resolutions of 2.15 and 2.60 A, respectively.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3W98
 
 | | Crystal Structure of Human Nucleosome Core Particle lacking H3.1 N-terminal region | | Descriptor: | 146-mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Iwasaki, W, Miya, Y, Horikoshi, N, Osakabe, A, Tachiwana, H, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2013-04-01 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Contribution of histone N-terminal tails to the structure and stability of nucleosomes
FEBS Open Bio, 3, 2013
|
|
1PI7
 
 | | Structure of the channel-forming trans-membrane domain of Virus protein "u" (Vpu) from HIV-1 | | Descriptor: | VPU protein | | Authors: | Park, S.H, Mrse, A.A, Nevzorov, A.A, Mesleh, M.F, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2003-05-29 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Three-dimensional structure of the channel-forming trans-membrane domain of virus protein "u" (Vpu) from HIV-1
J.Mol.Biol., 333, 2003
|
|
2B00
 
 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Glycocholate | | Descriptor: | CALCIUM ION, GLYCOCHOLIC ACID, Phospholipase A2, ... | | Authors: | Pan, Y.H, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2005-09-12 | | Release date: | 2006-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for bile salt inhibition of pancreatic phospholipase A2.
J.Mol.Biol., 369, 2007
|
|
2B04
 
 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Glycochenodeoxycholate | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCOCHENODEOXYCHOLIC ACID, ... | | Authors: | Pan, Y.H, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2005-09-12 | | Release date: | 2006-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for bile salt inhibition of pancreatic phospholipase A2.
J.Mol.Biol., 369, 2007
|
|
3UF1
 
 | | Crystal Structure of Vimentin (fragment 144-251) from Homo sapiens, Northeast Structural Genomics Consortium Target HR4796B | | Descriptor: | Vimentin | | Authors: | Kuzin, A, Abashidze, M, Vorobiev, S.M, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-30 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and X-ray crystallography.
J.Biol.Chem., 287, 2012
|
|
2B0Y
 
 | | Solution Structure of a peptide mimetic of the fourth cytoplasmic loop of the G-protein coupled CB1 cannabinoid receptor | | Descriptor: | Cannabinoid receptor 1 | | Authors: | Grace, C.R, Cowsik, S.M, Shim, J.Y, Welsh, W.J, Howlett, A.C. | | Deposit date: | 2005-09-15 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Unique helical conformation of the fourth cytoplasmic loop of the CB1 cannabinoid receptor in a negatively charged environment.
J.Struct.Biol., 159, 2007
|
|
4HB0
 
 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(K541Q,K542Q,R543S,K545Q,K548Q,K579Q)-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3A6N
 
 | | The nucleosome containing a testis-specific histone variant, human H3T | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Tachiwana, H, Kagawa, W, Osakabe, A, Koichiro, K, Shiga, T, Kimura, H, Kurumizaka, H. | | Deposit date: | 2009-09-04 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2BBO
 
 | | Human NBD1 with Phe508 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Kearins, M.C, Conners, K, Zhao, X, Lu, F, Sauder, J.M, Emtage, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry.
J.Mol.Biol., 396, 2010
|
|
4HAX
 
 | |
2AZY
 
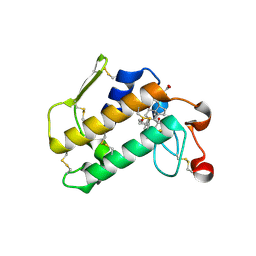 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with Cholate | | Descriptor: | CALCIUM ION, CHOLIC ACID, Phospholipase A2, ... | | Authors: | Pan, Y.H, Bahnson, B.J, Jain, M.K. | | Deposit date: | 2005-09-12 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for bile salt inhibition of pancreatic phospholipase A2.
J.Mol.Biol., 369, 2007
|
|
1XFA
 
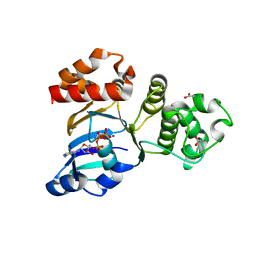 | | Structure of NBD1 from murine CFTR- F508R mutant | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Thibodeau, P.H, Brautigam, C.A, Machius, M, Thomas, P.J. | | Deposit date: | 2004-09-14 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Side chain and backbone contributions of Phe508 to CFTR folding.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2B11
 
 | |
1G5Q
 
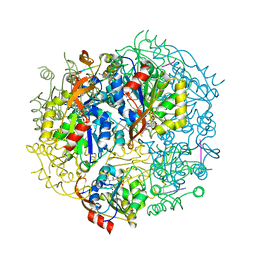 | | EPID H67N COMPLEXED WITH SUBSTRATE PEPTIDE DSYTC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, EPIDERMIN MODIFYING ENZYME EPID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbacher, S. | | Deposit date: | 2000-11-02 | | Release date: | 2001-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of the peptidyl-cysteine decarboxylase EpiD complexed with a pentapeptide substrate.
EMBO J., 19, 2000
|
|
1XP0
 
 | | Catalytic Domain Of Human Phosphodiesterase 5A In Complex With Vardenafil | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-07 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
4HAV
 
 | |
1GIP
 
 | |
4HB2
 
 | | Crystal structure of CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H8S
 
 | | Crystal structure of human APPL2BARPH domain | | Descriptor: | DCC-interacting protein 13-beta | | Authors: | Martin, J.L, King, G.J. | | Deposit date: | 2012-09-23 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Membrane Curvature Protein Exhibits Interdomain Flexibility and Binds a Small GTPase.
J.Biol.Chem., 287, 2012
|
|
4HFV
 
 | | Crystal structure of lpg1851 protein from Legionella pneumophila (putative T4SS effector) | | Descriptor: | CITRIC ACID, SUCCINIC ACID, Uncharacterized protein | | Authors: | Michalska, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-05 | | Release date: | 2012-11-07 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
4HAU
 
 | |
1PHK
 
 | | TWO STRUCTURES OF THE CATALYTIC DOMAIN OF PHOSPHORYLASE, KINASE: AN ACTIVE PROTEIN KINASE COMPLEXED WITH NUCLEOTIDE, SUBSTRATE-ANALOGUE AND PRODUCT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE | | Authors: | Owen, D.J, Noble, M.E.M, Garman, E.F, Papageorgiou, A.C, Johnson, L.N. | | Deposit date: | 1996-03-15 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two structures of the catalytic domain of phosphorylase kinase: an active protein kinase complexed with substrate analogue and product.
Structure, 3, 1995
|
|
