1PPB
 
 | |
1PPH
 
 | | GEOMETRY OF BINDING OF THE NALPHA-TOSYLATED PIPERIDIDES OF M-AMIDINO-, P-AMIDINO-AND P-GUANIDINO PHENYLALANINE TO THROMBIN AND TRYPSIN: X-RAY CRYSTAL STRUCTURES OF THEIR TRYPSIN COMPLEXES AND MODELING OF THEIR THROMBIN COMPLEXES | | Descriptor: | 3-[(2S)-2-{[(4-methylphenyl)sulfonyl]amino}-3-oxo-3-piperidin-1-ylpropyl]benzenecarboximidamide, CALCIUM ION, SULFATE ION, ... | | Authors: | Bode, W, Turk, D. | | Deposit date: | 1991-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Geometry of binding of the N alpha-tosylated piperidides of m-amidino-, p-amidino- and p-guanidino phenylalanine to thrombin and trypsin. X-ray crystal structures of their trypsin complexes and modeling of their thrombin complexes.
FEBS Lett., 287, 1991
|
|
1PVB
 
 | | X-RAY STRUCTURE OF A NEW CRYSTAL FORM OF PIKE 4.10 PARVALBUMIN | | Descriptor: | AMMONIUM ION, CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J. | | Deposit date: | 1995-01-05 | | Release date: | 1995-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray structure of a new crystal form of pike 4.10 beta parvalbumin.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1PAG
 
 | | THE 2.5 ANGSTROMS STRUCTURE OF POKEWEED ANTIVIRAL PROTEIN | | Descriptor: | FORMYCIN-5'-MONOPHOSPHATE, POKEWEED ANTIVIRAL PROTEIN | | Authors: | Monzingo, A.F, Collins, E.J, Ernst, S.R, Irvin, J.D, Robertus, J.D. | | Deposit date: | 1992-10-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.5 A structure of pokeweed antiviral protein.
J.Mol.Biol., 233, 1993
|
|
1PDZ
 
 | | X-RAY STRUCTURE AND CATALYTIC MECHANISM OF LOBSTER ENOLASE | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, ENOLASE, MANGANESE (II) ION | | Authors: | Janin, J, Duquerroy, S, Camus, C, Le Bras, G. | | Deposit date: | 1995-06-05 | | Release date: | 1995-11-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure and catalytic mechanism of lobster enolase.
Biochemistry, 34, 1995
|
|
1PLG
 
 | | EVIDENCE FOR THE EXTENDED HELICAL NATURE OF POLYSACCHARIDE EPITOPES. THE 2.8 ANGSTROMS RESOLUTION STRUCTURE AND THERMODYNAMICS OF LIGAND BINDING OF AN ANTIGEN BINDING FRAGMENT SPECIFIC FOR ALPHA-(2->8)-POLYSIALIC ACID | | Descriptor: | IGG2A=KAPPA= | | Authors: | Evans, S.V, Sigurskjold, B.W, Jennings, H.J, Brisson, J.-R, Tse, W.C, To, R, Altman, E, Frosch, M, Weisgerber, C, Kratzin, H, Klebert, S, Vaesen, M, Bitter-Suermann, D, Rose, D.R, Young, N.M, Bundle, D.R. | | Deposit date: | 1995-04-24 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for the extended helical nature of polysaccharide epitopes. The 2.8 A resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for alpha-(2-->8)-polysialic acid.
Biochemistry, 34, 1995
|
|
1PVA
 
 | | COMPARISON BETWEEN THE CRYSTAL AND THE SOLUTION STRUCTURES OF THE EF HAND PARVALBUMIN (ALPHA COMPONENT FROM PIKE MUSCLE) | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Roquet, F, Rambaud, J, Parello, J. | | Deposit date: | 1995-01-16 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Comparison between the Crystal and the Solution Structures of the EF Hand Parvalbumin (Alpha Component from Pike Muscle)
To be Published
|
|
1PLR
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
1PRT
 
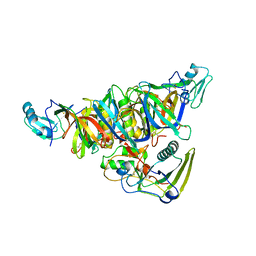 | | THE CRYSTAL STRUCTURE OF PERTUSSIS TOXIN | | Descriptor: | PERTUSSIS TOXIN (SUBUNIT S1), PERTUSSIS TOXIN (SUBUNIT S2), PERTUSSIS TOXIN (SUBUNIT S3), ... | | Authors: | Stein, P.E, Read, R.J. | | Deposit date: | 1993-11-22 | | Release date: | 1995-01-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of pertussis toxin.
Structure, 2, 1994
|
|
1POE
 
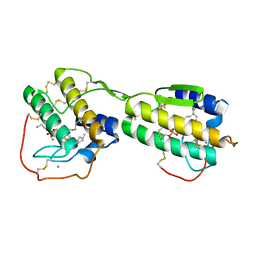 | |
1PVC
 
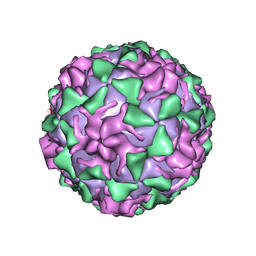 | |
1PSH
 
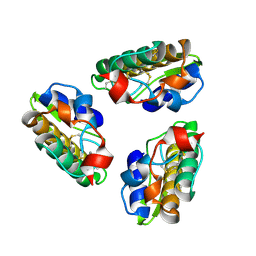 | |
1H87
 
 | | Gadolinium derivative of tetragonal Hen Egg-White Lysozyme at 1.7 A resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Girard, E, Chantalat, L, Vicat, J, Kahn, R. | | Deposit date: | 2001-01-25 | | Release date: | 2002-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Gd-Hp-Do3A, a Complex to Obtain High-Phasing-Power Heavy Atom Derivatives for Sad and MAD Experiments. Results with Tetragonal Hen Egg-White Lysozyme
Acta Crystallogr.,Sect.D, 58, 2001
|
|
1HCU
 
 | | alpha-1,2-mannosidase from Trichoderma reesei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-1,2-MANNOSIDASE, CALCIUM ION | | Authors: | Van Petegem, F, Contreras, H, Contreras, R, Van Beeumen, J. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Trichoderma Reesei Alpha-1,2-Mannosidase: Structural Basis for the Cleavage of Four Consecutive Mannose Residues
J.Mol.Biol., 312, 2001
|
|
1QOL
 
 | | STRUCTURE OF THE FMDV LEADER PROTEASE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PROTEASE (NONSTRUCTURAL PROTEIN P20A) | | Authors: | Guarne, A, Tormo, J, Kirchweger, R, Pfistermueller, D, Skern, T, Fita, I. | | Deposit date: | 1999-11-13 | | Release date: | 2000-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Foot-and-Mouth Disease Virus Leader Protease: A Papain-Like Fold Adapted for Self-Processing and Eif4G Recognition.
Embo J., 17, 1998
|
|
1QAG
 
 | | Actin binding region of the dystrophin homologue utrophin | | Descriptor: | UTROPHIN ACTIN BINDING REGION | | Authors: | Keep, N.H, Winder, S.J, Moores, C.A, Walke, S, Norwood, F.L.M, Kendrick-Jones, J. | | Deposit date: | 1999-03-05 | | Release date: | 2000-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the actin-binding region of utrophin reveals a head-to-tail dimer
Structure Fold.Des., 7, 1999
|
|
1H7U
 
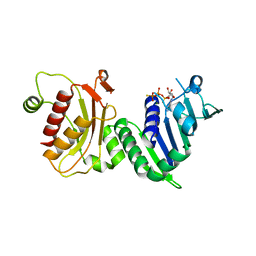 | | hPMS2-ATPgS | | Descriptor: | MAGNESIUM ION, MISMATCH REPAIR ENDONUCLEASE PMS2, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
1QFG
 
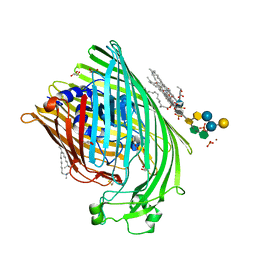 | | E. COLI FERRIC HYDROXAMATE RECEPTOR (FHUA) | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Welte, W, Hofmann, E, Lindner, B, Holst, O, Coulton, J.W, Diederichs, K. | | Deposit date: | 1999-04-10 | | Release date: | 2000-07-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins.
Structure Fold.Des., 8, 2000
|
|
1QE6
 
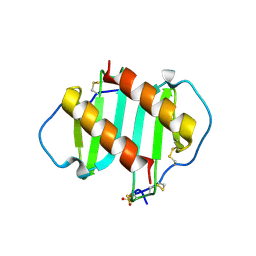 | | INTERLEUKIN-8 WITH AN ADDED DISULFIDE BETWEEN RESIDUES 5 AND 33 (L5C/H33C) | | Descriptor: | INTERLEUKIN-8 VARIANT, SULFATE ION | | Authors: | Gerber, N, Lowman, H, Artis, D.R, Eigenbrot, C. | | Deposit date: | 1999-07-13 | | Release date: | 2000-03-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Receptor-binding conformation of the "ELR" motif of IL-8: X-ray structure of the L5C/H33C variant at 2.35 A resolution.
Proteins, 38, 2000
|
|
1QGR
 
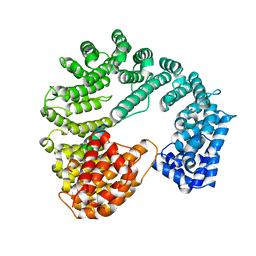 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA (II CRYSTAL FORM, GROWN AT LOW PH) | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-05-04 | | Release date: | 1999-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
1QFY
 
 | | PEA FNR Y308S MUTANT IN COMPLEX WITH NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (FERREDOXIN: NADP+ REDUCTASE), ... | | Authors: | Deng, Z, Aliverti, A, Zanetti, G, Arakaki, A.K, Ottado, J, Orellano, E.G, Calcaterra, N.B, Ceccarelli, E.A, Carrillo, N, Karplus, P.A. | | Deposit date: | 1999-04-18 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A productive NADP+ binding mode of ferredoxin-NADP + reductase revealed by protein engineering and crystallographic studies.
Nat.Struct.Biol., 6, 1999
|
|
1OI4
 
 | |
1OXA
 
 | | CYTOCHROME P450 (DONOR:O2 OXIDOREDUCTASE) | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, CYTOCHROME P450 ERYF, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cupp-Vickery, J.R, Poulos, T.L. | | Deposit date: | 1995-07-14 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of cytochrome P450eryF involved in erythromycin biosynthesis.
Nat.Struct.Biol., 2, 1995
|
|
1OKL
 
 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKL INHIBITOR 5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONAMIDE | | Descriptor: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Elbaum, D, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected binding mode of the sulfonamide fluorophore 5-dimethylamino-1-naphthalene sulfonamide to human carbonic anhydrase II. Implications for the development of a zinc biosensor.
J.Biol.Chem., 271, 1996
|
|
1ONR
 
 | | STRUCTURE OF TRANSALDOLASE B | | Descriptor: | TRANSALDOLASE B | | Authors: | Jia, J, Huang, W, Lindqvist, Y, Schneider, G. | | Deposit date: | 1996-08-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of transaldolase B from Escherichia coli suggests a circular permutation of the alpha/beta barrel within the class I aldolase family.
Structure, 4, 1996
|
|
