4UZX
 
 | | High-resolution NMR structures of the domains of Saccharomyces cerevisiae Tho1 | | Descriptor: | PROTEIN THO1 | | Authors: | Jacobsen, J.O.B, Allen, M.D, Freund, S.M.V, Bycroft, M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution NMR Structures of the Domains of Saccharomyces Cerevisiae Tho1.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4OUR
 
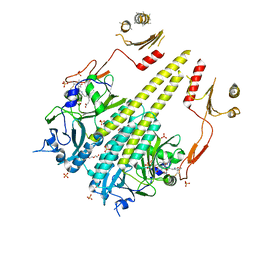 | | Crystal structure of Arabidopsis thaliana phytochrome B photosensory module | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, Phytochrome B, ... | | Authors: | Sethe Burgie, E, Bussell, A.N, Walker, J.M, Dubiel, K, Vierstra, R.D. | | Deposit date: | 2014-02-18 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3L78
 
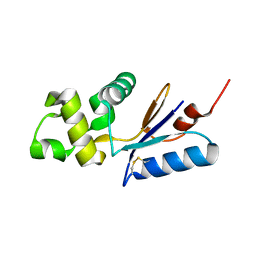 | |
4W5V
 
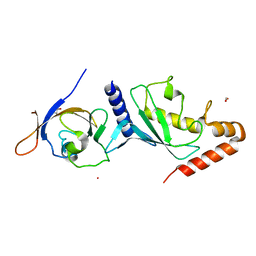 | | Crystal structure of Human SUMO E2-conjugating enzyme (Ubc9) in complex with E1-activating enzyme (Uba2) ubiquitin fold domain (Ufd) | | Descriptor: | FORMIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Boucher, L.E, Reiter, K.H, Matunis, M.J, Bosch, J. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human SUMO complex
To Be Published
|
|
4WFZ
 
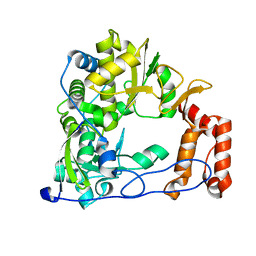 | |
4WID
 
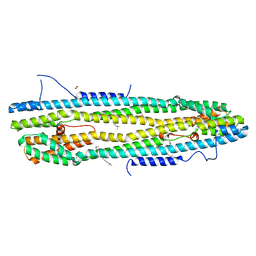 | | Crystal structure of the immediate-early 1 protein (IE1) at 2.31 angstrom (tetragonal form after crystal dehydration) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RhUL123 | | Authors: | Klingl, S, Scherer, M, Sevvana, M, Muller, Y.A, Stamminger, T. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Cytomegalovirus IE1 Protein Reveals Targeting of TRIM Family Member PML via Coiled-Coil Interactions.
Plos Pathog., 10, 2014
|
|
2DMT
 
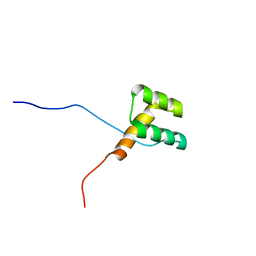 | | Solution structure of the homeobox domain of Homeobox protein BarH-like 1 | | Descriptor: | Homeobox protein BarH-like 1 | | Authors: | Ohnishi, S, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homeobox domain of Homeobox protein BarH-like 1
To be Published
|
|
8T4S
 
 | |
4WLA
 
 | |
4Q2U
 
 | |
4ORC
 
 | | Crystal structure of mammalian calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Ma, L, Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
4WFY
 
 | |
4WIC
 
 | | Immediate-early 1 protein (IE1) of rhesus macaque cytomegalovirus | | Descriptor: | RhUL123 | | Authors: | Klingl, S, Scherer, M, Sevvana, M, Muller, Y.A, Stamminger, T. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Controlled crystal dehydration triggers a space-group switch and shapes the tertiary structure of cytomegalovirus immediate-early 1 (IE1) protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4P43
 
 | |
4MLO
 
 | | 1.65A resolution structure of ToxT from Vibrio cholerae (P21 Form) | | Descriptor: | CHLORIDE ION, PALMITOLEIC ACID, TCP pilus virulence regulatory protein | | Authors: | Lovell, S, Wehmeyer, G, Battaile, K.P, Li, J, Egan, S. | | Deposit date: | 2013-09-06 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1.65 angstrom resolution structure of the AraC-family transcriptional activator ToxT from Vibrio cholerae.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
4P20
 
 | | Crystal structures of the bacterial ribosomal decoding site complexed with amikacin | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, 5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Kondo, J, Francois, B, Russell, R.J.M, Murray, J.B, Westhof, E. | | Deposit date: | 2014-02-28 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the bacterial ribosomal decoding site complexed with amikacin containing the gamma-amino-alpha-hydroxybutyryl (haba) group.
Biochimie, 88, 2006
|
|
8TWD
 
 | | Structure of IraM-bound RssB | | Descriptor: | ACETATE ION, Anti-adapter protein IraM, Regulator of RpoS | | Authors: | Brugger, C, Deaconescu, A.M. | | Deposit date: | 2023-08-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | IraM remodels the RssB segmented helical linker to stabilize sigma s against degradation by ClpXP.
J.Biol.Chem., 300, 2023
|
|
7M2F
 
 | | CDK2 with compound 14 inhibitor with carboxylate | | Descriptor: | Cyclin-dependent kinase 2, [(1r,4r)-4-{4-[4-(5-fluoro-2-methoxyphenyl)-1H-pyrrolo[2,3-b]pyridin-2-yl]-3,6-dihydropyridin-1(2H)-yl}cyclohexyl]acetic acid | | Authors: | Longenecker, K.L, Qiu, W, Korepanova, A, Tong, Y. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Balancing Properties with Carboxylates: A Lead Optimization Campaign for Selective and Orally Active CDK9 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
3LYA
 
 | |
8T85
 
 | | Structure of RssB bound to beryllofluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Brugger, C, Schwartz, J, Deaconescu, A.M. | | Deposit date: | 2023-06-21 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of phosphorylated-like RssB, the adaptor delivering sigma s to the ClpXP proteolytic machinery, reveals an interface switch for activation.
J.Biol.Chem., 299, 2023
|
|
4DUG
 
 | | Crystal Structure of Circadian Clock Protein KaiC E318A Mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION, ... | | Authors: | Egli, M, Mori, T, Pattanayek, R, Xu, Y, Qin, X, Johnson, C.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.292 Å) | | Cite: | Dephosphorylation of the Core Clock Protein KaiC in the Cyanobacterial KaiABC Circadian Oscillator Proceeds via an ATP Synthase Mechanism.
Biochemistry, 51, 2012
|
|
2UYE
 
 | | Double mutant Y110S,F111V DntR from Burkholderia sp. strain DNT in complex with thiocyanate | | Descriptor: | GLYCEROL, REGULATORY PROTEIN, THIOCYANATE ION | | Authors: | Lonneborg, R, Smirova, I, Dian, C, leonard, G.A, McSweeney, S, Brzezinski, P. | | Deposit date: | 2007-04-04 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In Vivo and in Vitro Investigation of Transcriptional Regulation by Dntr.
J.Mol.Biol., 372, 2007
|
|
2DMS
 
 | | Solution structure of the homeobox domain of Homeobox protein OTX2 | | Descriptor: | Homeobox protein OTX2 | | Authors: | Ohnishi, S, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homeobox domain of Homeobox protein OTX2
To be Published
|
|
8GME
 
 | |
7MFD
 
 | | Autoinhibited BRAF:(14-3-3)2:MEK complex with the BRAF RBD resolved | | Descriptor: | 14-3-3 protein zeta/delta, Dual specificity mitogen-activated protein kinase kinase 1, N-(3-fluoro-4-{[4-methyl-2-oxo-7-(pyrimidin-2-yloxy)-2H-chromen-3-yl]methyl}pyridin-2-yl)-N'-methylsulfuric diamide, ... | | Authors: | Martinez Fiesco, J.A, Ping, Z, Durrant, D.E, Morrison, D.K. | | Deposit date: | 2021-04-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the BRAF monomer-to-dimer transition mediated by RAS binding.
Nat Commun, 13, 2022
|
|
