7ZYV
 
 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.13 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, ... | | Authors: | Sarewicz, M, Szwalec, M, Pintscher, S, Indyka, P, Rawski, M, Pietras, R, Mielecki, B, Koziej, L, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-25 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
8XOQ
 
 | | Human Calcium and Integrin Binding Protein 2 (CIB2) Fusion to TMC1 CBD-1 domain at 2.4 Angstroms resolution | | Descriptor: | MAGNESIUM ION, Transmembrane channel-like protein 1,Calcium and integrin-binding family member 2 | | Authors: | Li, Y.H, Chen, J.S, Zhang, X, Wang, C. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural insights into calcium-dependent CIB2-TMC1 interaction in hair cell mechanotransduction.
Commun Biol, 8, 2025
|
|
9BQX
 
 | |
4HU1
 
 | | Crystal structure of human carbonic anhydrase isozyme XIII with the inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 13, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Substituted-2,3,5,6-tetrafluorobenzenesulfonamides as inhibitors of carbonic anhydrases I, II, VII, XII, and XIII.
Bioorg.Med.Chem., 21, 2013
|
|
8TG3
 
 | | tRNA 2'-phosphotransferase (Tpt1) from Aeropyrum pernix in complex with ADP-ribose-1" -phosphate | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NITRATE ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2023-07-12 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for Tpt1-catalyzed 2'-PO 4 transfer from RNA and NADP(H) to NAD.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HBF
 
 | |
7MST
 
 | | Phosphorylated human E105Qa GTP-specific succinyl-CoA synthetase complexed with coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, GLYCEROL, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2021-05-12 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The structure of succinyl-CoA synthetase bound to the succinyl-phosphate intermediate clarifies the catalytic mechanism of ATP-citrate lyase
Acta Crystallogr.,Sect.F, 78, 2022
|
|
5GWX
 
 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with S-adenosylmethionine and sarcosine | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-09-14 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity.
Sci Rep, 6, 2016
|
|
8T1Q
 
 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8A6F
 
 | | Small molecule stabilizer (compound 8) for C-RAF and 14-3-3 | | Descriptor: | 1-[4-[4-chloranyl-3-(trifluoromethyl)phenyl]-4-oxidanyl-piperidin-1-yl]-3-[2-(dimethylamino)ethyldisulfanyl]propan-1-one, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
7MSR
 
 | | Human E105Qa GTP-specific succinyl-CoA synthetase complexed with succinyl-phosphate, magnesium ion and desulfo-coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, 4-oxo-4-(phosphonooxy)butanoic acid, DESULFO-COENZYME A, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2021-05-12 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The structure of succinyl-CoA synthetase bound to the succinyl-phosphate intermediate clarifies the catalytic mechanism of ATP-citrate lyase
Acta Crystallogr.,Sect.F, 78, 2022
|
|
5KO5
 
 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with cytosine | | Descriptor: | 1,2-ETHANEDIOL, 6-AMINOPYRIMIDIN-2(1H)-ONE, Purine nucleoside phosphorylase | | Authors: | Torini, J.R, Romanello, L, Bird, L, Owens, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.
PLoS ONE, 13, 2018
|
|
7TGT
 
 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor A26 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-7-benzyl-4-[(furan-2-yl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
6ARH
 
 | |
8SR2
 
 | | particulate methane monooxygenase incubated with 4,4,4-trifluorobutanol | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
9EW4
 
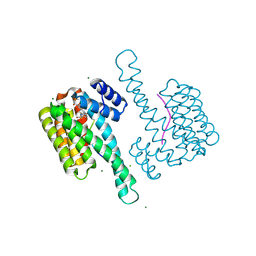 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide 12mer (pS259) and compound 86 (1124384) | | Descriptor: | 1-[(5~{R})-2-(4-bromanyl-3-fluoranyl-phenyl)sulfonyl-2,7-diazaspiro[4.4]nonan-7-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2024-04-03 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
5NRW
 
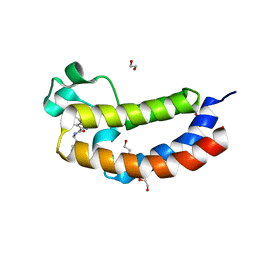 | |
9EW3
 
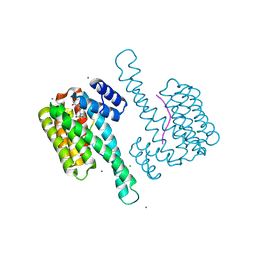 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide 12-mer (pS259) and compound 78 (1084378) | | Descriptor: | 1-[8-(4-bromophenyl)sulfonyl-5-oxa-2,8-diazaspiro[3.5]nonan-2-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2024-04-03 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
7QQG
 
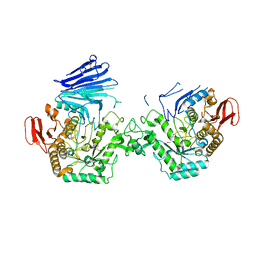 | | Crystal structure of MYORG bound to 1-deoxygalactonojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The primary familial brain calcification-associated protein MYORG is an alpha-galactosidase with restricted substrate specificity.
Plos Biol., 20, 2022
|
|
5KKC
 
 | | l-lactate dehydrogenase from rabbit muscle with the inhibitor 6DHNAD | | Descriptor: | L-lactate dehydrogenase A chain, SULFATE ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(5-aminocarbonyl-2~{H}-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Meneely, K.M, Moran, G.R, Lamb, A.L. | | Deposit date: | 2016-06-21 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Ligand binding phenomena that pertain to the metabolic function of renalase.
Arch.Biochem.Biophys., 612, 2016
|
|
7QQH
 
 | | Crystal structure of MYORG (D520N) in complex with Gal-a1,4-Glc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The primary familial brain calcification-associated protein MYORG is an alpha-galactosidase with restricted substrate specificity.
Plos Biol., 20, 2022
|
|
6WLE
 
 | | Crystal structure of the Zeitlupe light-state mimic G46A | | Descriptor: | 1,2-ETHANEDIOL, Adagio protein 1, FLAVIN MONONUCLEOTIDE | | Authors: | Zoltowski, B, Green, R. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Steric and Electronic Interactions at Gln154 in ZEITLUPE Induce Reorganization of the LOV Domain Dimer Interface.
Biochemistry, 60, 2021
|
|
6WLP
 
 | | Crystal Structure of the ZTL light-state mimic G46S | | Descriptor: | 1,2-ETHANEDIOL, Adagio protein 1, FLAVIN MONONUCLEOTIDE | | Authors: | Zoltowski, B, Green, R. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Steric and Electronic Interactions at Gln154 in ZEITLUPE Induce Reorganization of the LOV Domain Dimer Interface.
Biochemistry, 60, 2021
|
|
5G1W
 
 | | Apo Structure of Linalool Dehydratase-Isomerase | | Descriptor: | 1,2-ETHANEDIOL, LINALOOL DEHYDRATASE/ISOMERASE, METHYLMALONIC ACID | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
8QQL
 
 | |
