7K9X
 
 | |
1FKM
 
 | | CRYSTAL STRUCTURE OF THE YPT/RAB-GAP DOMAIN OF GYP1P | | Descriptor: | PROTEIN (GYP1P) | | Authors: | Rak, A, Fedorov, R, Alexandrov, K, Albert, S, Goody, R.S, Gallwitz, D, Scheidig, A.J. | | Deposit date: | 2000-08-09 | | Release date: | 2001-02-09 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the GAP domain of Gyp1p: first insights into interaction with Ypt/Rab proteins.
EMBO J., 19, 2000
|
|
7K3V
 
 | | Apoferritin structure at 1.34 angstrom resolution determined from a 300 kV Titan Krios G3i electron microscope with K3 detector | | Descriptor: | Ferritin heavy chain, SODIUM ION, ZINC ION | | Authors: | Zhang, K, Pintilie, G, Li, S, Schmid, M, Chiu, W. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.34 Å) | | Cite: | Resolving individual atoms of protein complex by cryo-electron microscopy.
Cell Res., 30, 2020
|
|
1FA3
 
 | |
1FGI
 
 | |
1MB0
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 8.0 IN COMPLEX WITH MN2+ | | Descriptor: | MANGANESE (II) ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
1MMT
 
 | | Crystal structure of ternary complex of the catalytic domain of human phenylalanine hydroxylase (Fe(II)) complexed with tetrahydrobiopterin and norleucine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, FE (II) ION, NORLEUCINE, ... | | Authors: | Andersen, O.A, Flatmark, T, Hough, E. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A resolution crystal structures of the ternary complexes of human phenylalanine hydroxylase catalytic domain with tetrahydrobiopterin and 3-(2-thienyl)-L-alanine or L-norleucine: substrate specificity and molecular motions related to substrate binding
J.Mol.Biol., 333, 2003
|
|
6BX6
 
 | | AMP-Activated protein kinase (AMPK) inhibition by SBI-0206965: alpha 2 kinase domain bound to SBI-0206965 | | Descriptor: | 2-({5-bromo-2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}oxy)-N-methylbenzene-1-carboximidic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-2 | | Authors: | Dite, T.A, Langendorf, C.G, Scott, J.W, Oakhill, J.S. | | Deposit date: | 2017-12-17 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AMP-activated protein kinase selectively inhibited by the type II inhibitor SBI-0206965.
J. Biol. Chem., 293, 2018
|
|
6BXP
 
 | |
1MCB
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCI
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-D-PHE-L-HIS-D-PRO-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
7JSG
 
 | | Adeno-Associated Virus 2 Rep68 HD-Heptamer-ssDNA with ATPgS | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
6BQD
 
 | | TAF1-BD2 bromodomain in complex with (E)-3-(6-(but-2-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide | | Descriptor: | 3-{6-[(2E)-but-2-en-1-yl]-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl}-N,N-dimethylbenzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.136 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
1MCS
 
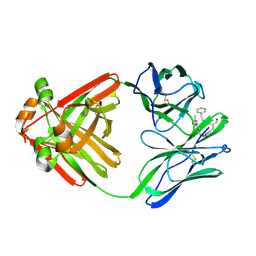 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
7K11
 
 | |
6BSC
 
 | |
1FLR
 
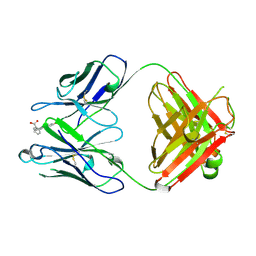 | | 4-4-20 FAB FRAGMENT | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, 4-4-20 (IG*G2A=KAPPA=) FAB FRAGMENT | | Authors: | Whitlow, M. | | Deposit date: | 1995-01-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 A structure of anti-fluorescein 4-4-20 Fab.
Protein Eng., 8, 1995
|
|
6BYH
 
 | |
1MQ2
 
 | | Human DNA Polymerase Beta Complexed With Gapped DNA Containing an 8-oxo-7,8-dihydro-Guanine and dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*(8OG)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(2DA))-3', ... | | Authors: | Krahn, J.M, Beard, W.A, Miller, H, Grollman, A.P, Wilson, S.H. | | Deposit date: | 2002-09-13 | | Release date: | 2003-01-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of DNA Polymerase beta with the Mutagenic DNA Lesion 8-oxodeoxyguanine Reveals
Structural Insights into its Coding Potential
Structure, 11, 2003
|
|
6BZT
 
 | | Crystal structure of halogenase PltM L111Y mutant in complex with FAD | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pang, A.H, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2017-12-26 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unusual substrate and halide versatility of phenolic halogenase PltM.
Nat Commun, 10, 2019
|
|
1KVR
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
7JVK
 
 | |
7K10
 
 | |
1KVS
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
6C18
 
 | | FGFR1 kinase complex with inhibitor SN37115 | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{(3S)-1-[(2E)-4-(dimethylamino)but-2-enoyl]pyrrolidin-3-yl}-7-[(propan-2-yl)amino]-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Understanding the structural requirements for covalent inhibition of FGFR1-3
To Be Published
|
|
