5JTR
 
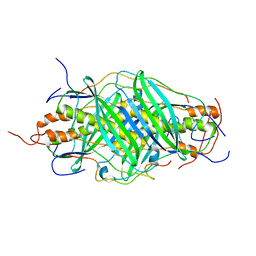 | | The structure of chaperone SecB in complex with unstructured MBP binding site e | | Descriptor: | Maltose-binding periplasmic protein, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JS7
 
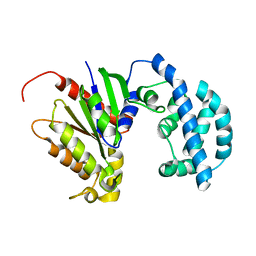 | | Structural model of a apo G-protein alpha subunit determined with NMR residual dipolar couplings and SAXS | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1 | | Authors: | Goricanec, D, Stehle, R, Grigoriu, S, Wagner, G, Hagn, F. | | Deposit date: | 2016-05-07 | | Release date: | 2016-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JS8
 
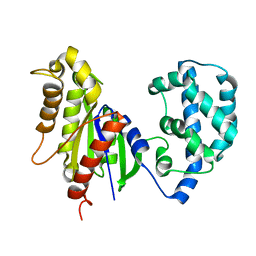 | | Structural Model of a Protein alpha subunit in complex with GDP obtained with SAXS and NMR residual couplings | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1 | | Authors: | Goricanec, D, Stehle, R, Grigoriu, S, Wagner, G, Hagn, F. | | Deposit date: | 2016-05-07 | | Release date: | 2016-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1H76
 
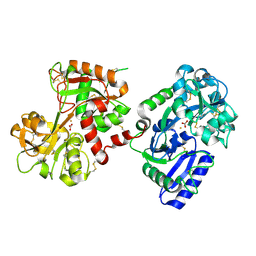 | | The crystal structure of diferric porcine serum transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Hall, D.R, Hadden, J.M, Leonard, G.A, Bailey, S, Neu, M, Winn, M, Lindley, P.F. | | Deposit date: | 2001-07-03 | | Release date: | 2002-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal and Molecular Structures of Diferric Porcine and Rabbit Serum Transferrins at Resolutions of 2.15 And 2.60A, Respectively
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2WXB
 
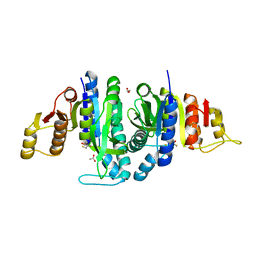 | | Acetylglutamate kinase from Escherichia coli free of substrates | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Ramon-Maiques, S, Gil-Ortiz, F, Rubio, V. | | Deposit date: | 2009-11-06 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Crystal Structures of Escherichia Coli N-Acetyl-L-Glutamate Kinase Demonstrate the Cycling between Open and Closed Conformations.
J.Mol.Biol., 399, 2010
|
|
5KK9
 
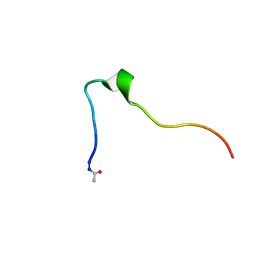 | | Connexin 32 G12R N-Terminal Mutant, | | Descriptor: | Gap junction beta-1 protein | | Authors: | Dowd, T.L, Barigello, T.A. | | Deposit date: | 2016-06-21 | | Release date: | 2016-09-28 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Structural studies of N-terminal mutants of Connexin 26 and Connexin 32 using (1)H NMR spectroscopy.
Arch.Biochem.Biophys., 608, 2016
|
|
1H7Y
 
 | | Translationally Controlled Tumor-associated Protein p23fyp from Schizosaccharomyces pombe | | Descriptor: | TRANSLATIONALLY CONTROLLED TUMOR PROTEIN | | Authors: | Thaw, P, Baxter, N.J, Sedelnikova, S.E, Price, C, Waltho, J.P, Craven, C.J. | | Deposit date: | 2001-01-19 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones.
Nat. Struct. Biol., 8, 2001
|
|
1VCC
 
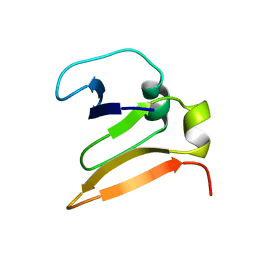 | |
3LTM
 
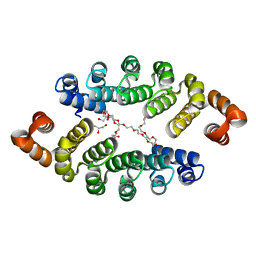 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | Descriptor: | Alpha-Rep4, DODECAETHYLENE GLYCOL, GLYCEROL, ... | | Authors: | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | Deposit date: | 2010-02-16 | | Release date: | 2010-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
3LLL
 
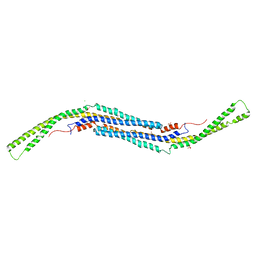 | | Crystal structure of mouse pacsin2 F-BAR domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, Protein kinase C and casein kinase substrate in neurons protein 2, ... | | Authors: | Rudolph, M.G. | | Deposit date: | 2010-01-29 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A hinge in the distal end of the PACSIN 2 F-BAR domain may contribute to membrane-curvature sensing.
J.Mol.Biol., 400, 2010
|
|
2X00
 
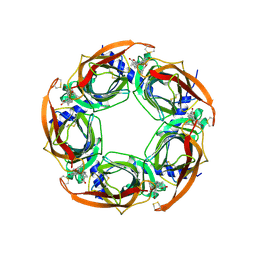 | | CRYSTAL STRUCTURE OF A-ACHBP IN COMPLEX WITH GYMNODIMINE A | | Descriptor: | GYMNODIMINE A, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Bourne, Y, Radic, Z, Araoz, R, Talley, T.T, Benoit, E, Servent, D, Taylor, P, Molgo, J, Marchot, P. | | Deposit date: | 2009-12-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Determinants in Phycotoxins and Achbp Conferring High Affinity Binding and Nicotinic Achr Antagonism.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5MTI
 
 | | Bamb_5917 Acyl-Carrier Protein | | Descriptor: | Phosphopantetheine-binding protein | | Authors: | Gallo, A, Kosol, S, Griffiths, D, Masschelein, J, Alkhalaf, L, Smith, H, Valentic, T, Tsai, S, Challis, G, Lewandowski, J.R. | | Deposit date: | 2017-01-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for chain release from the enacyloxin polyketide synthase
Nat.Chem., 2019
|
|
5MQX
 
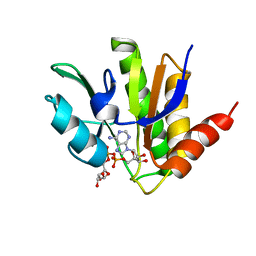 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus(VEEV) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein3 | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Matsoukas, M.T, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-12-21 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
5ML1
 
 | | NMR Structure of the Littorina littorea metallothionein, a snail MT folding into three distinct domains | | Descriptor: | CADMIUM ION, Putative metallothionein | | Authors: | Baumann, C, Beil, A, Jurt, S, Zerbe, O. | | Deposit date: | 2016-12-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Adaptation of a Protein to Increased Metal Stress: NMR Structure of a Marine Snail Metallothionein with an Additional Domain.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JC7
 
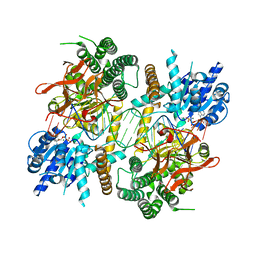 | |
5MN3
 
 | |
5JDT
 
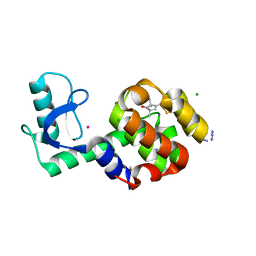 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at 100K | | Descriptor: | AZIDE ION, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Loll, B, Consentius, P, Gohlke, U, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2016-04-17 | | Release date: | 2016-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
4DHF
 
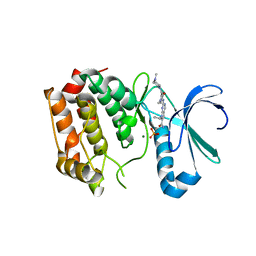 | | Structure of Aurora A mutant bound to Biogenidec cpd 15 | | Descriptor: | 7-cyclopentyl-2-({1-methyl-5-[(4-methylpiperazin-1-yl)carbonyl]-1H-pyrrol-3-yl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Silvian, L, Marcotte, D.J. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of 2,6,7-trisubstituted-7H-pyrrolo[2,3-d]pyrimidines as Aurora kinases inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2X0G
 
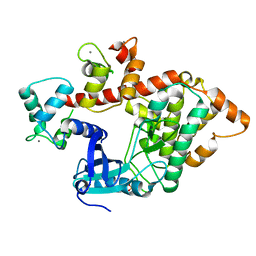 | | X-RAY STRUCTURE OF A DAP-KINASE CALMODULIN COMPLEX | | Descriptor: | CALCIUM ION, CALMODULIN, DEATH-ASSOCIATED PROTEIN KINASE 1, ... | | Authors: | Kuper, J, De Diego, I, Lehmann, F, Wilmanns, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis of the Death-Associated Protein Kinase-Calcium/Calmodulin Regulator Complex.
Sci.Signal, 3, 2010
|
|
1YSM
 
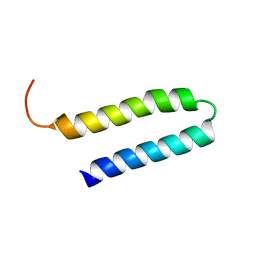 | | NMR Structure of N-terminal domain (Residues 1-77) of Siah-Interacting Protein. | | Descriptor: | Calcyclin-binding protein | | Authors: | Bhattacharya, S, Lee, Y.T, Michowski, W, Jastrzebska, B, Filipek, A, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2005-02-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Modular Structure of SIP Facilitates Its Role in Stabilizing Multiprotein Assemblies.
Biochemistry, 44, 2005
|
|
3SO0
 
 | |
5JAW
 
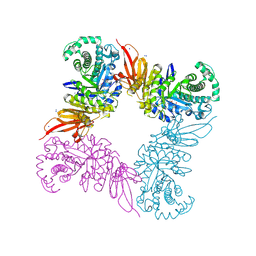 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | (1S,2S,3S,4S,5R,6R)-5-amino-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2016-04-12 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Towards broad spectrum activity-based glycosidase probes: synthesis and evaluation of deoxygenated cyclophellitol aziridines.
Chem. Commun. (Camb.), 53, 2017
|
|
3SBY
 
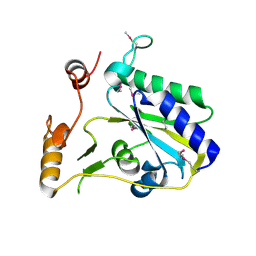 | | Crystal Structure of SeMet-Substituted Apo-MMACHC (1-244), a human B12 processing enzyme | | Descriptor: | Methylmalonic aciduria and homocystinuria type C protein | | Authors: | Koutmos, M, Gherasim, C, Smith, J.L, Banerjee, R. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural basis of multifunctionality in a vitamin B12-processing enzyme.
J.Biol.Chem., 286, 2011
|
|
2W6I
 
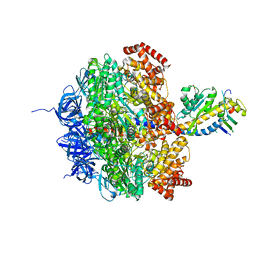 | | Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 4B. | | Descriptor: | ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT BETA, ... | | Authors: | Sanchez-Weatherby, J, Felisaz, F, Gobbo, A, Huet, J, Ravelli, R.B.G, Bowler, M.W, Cipriani, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Improving diffraction by humidity control: a novel device compatible with X-ray beamlines.
Acta Crystallogr. D Biol. Crystallogr., 65, 2009
|
|
3LAU
 
 | |
