7AON
 
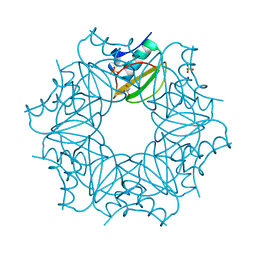 | | Crystal structure of CI2 double mutant L49I,I57V | | Descriptor: | GLYCEROL, SULFATE ION, Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Olsen, J.G, Teilum, K, Hamborg, L, Roche, J.V. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synergistic stabilization of a double mutant in chymotrypsin inhibitor 2 from a library screen in E. coli.
Commun Biol, 4, 2021
|
|
7AUD
 
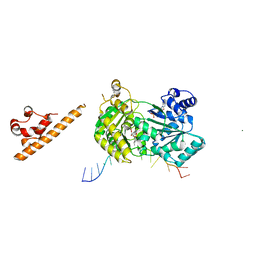 | |
7QO2
 
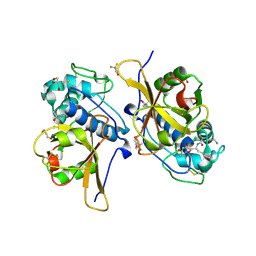 | | Peptide GAKSAA in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-12-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Peptide GAKSAA in complex with human cathepsin V C25A mutant
To Be Published
|
|
6CNP
 
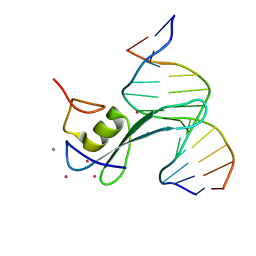 | |
6D0K
 
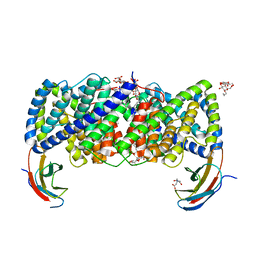 | | Crystal structure of a CLC-type fluoride/proton antiporter, E118Q mutant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7QW5
 
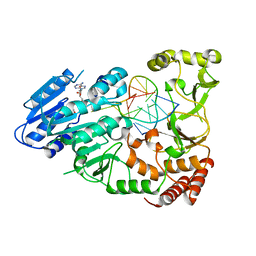 | | Adenine-specific DNA methyltransferase M.BseCI complexed with AdoHcy and cognate unmethylated DNA duplex | | Descriptor: | Modification methylase BseCI, S-ADENOSYL-L-HOMOCYSTEINE, Unmethylated DNA duplex | | Authors: | Mitsikas, D.A, Kouyianou, K, Kotsifaki, D, Providaki, M, Bouriotis, V, Glykos, N.M, Kokkinidis, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of M.BseCI DNA methyltransferase from Geobacillus stearothermophilus.
To Be Published
|
|
7B1A
 
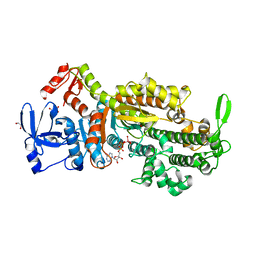 | | Myosin-II-AA mutant motor domain | | Descriptor: | 1,2-ETHANEDIOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Ewert, W, Preller, M. | | Deposit date: | 2020-11-24 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unraveling a Force-Generating Allosteric Pathway of Actomyosin Communication Associated with ADP and P i Release.
Int J Mol Sci, 22, 2020
|
|
6JD9
 
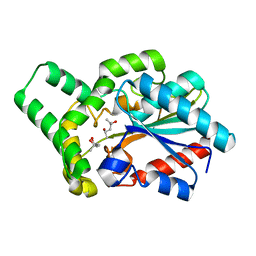 | | Proteus mirabilis lipase mutant - I118V/E130G | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase, CALCIUM ION | | Authors: | Heater, B.S, Chan, W.S, Chan, M.K. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Directed evolution of a genetically encoded immobilized lipase for the efficient production of biodiesel from waste cooking oil.
Biotechnol Biofuels, 12, 2019
|
|
7ZGL
 
 | |
7QQ2
 
 | |
7B33
 
 | | MST3 in complex with MRIA11 | | Descriptor: | 1,2-ETHANEDIOL, 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[4-[6-[bis(fluoranyl)methyl]pyridin-2-yl]-2-chloranyl-phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7ZHQ
 
 | | Crystal structure of TTBK1 in complex with compound 10 (7-009) | | Descriptor: | (3~{S})-1-(4-azanyl-3,5,12-triazatetracyclo[9.7.0.0^{2,7}.0^{13,18}]octadeca-1(11),2,4,6,13(18),14,16-heptaen-16-yl)-3-methyl-pent-1-yn-3-ol, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
7LTD
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 1 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7AGI
 
 | | The structure of Artemis variant H35D | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7ZHO
 
 | | Crystal structure of TTBK1 in complex with compound 3 (7-001) | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-(2-azanylpyrimidin-4-yl)-1~{H}-indol-5-yl]-2-methyl-but-3-yn-2-ol, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
6CPY
 
 | | Structure of apo GRMZM2G135359 pseudokinase | | Descriptor: | GRMZM2G135359 pseudokinase | | Authors: | Aquino, B, Counago, R.M, Godoi, P.H.C, Massirer, K.B, Elkins, J.M, Arruda, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-14 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of apo GRMZM2G135359 pseudokinase
To be Published
|
|
7ZJJ
 
 | | CspZ (BbCRASP-2) from Borrelia burgdorferi strain B379 | | Descriptor: | CspZ, NITRATE ION | | Authors: | Brangulis, K, Marcinkiewicz, A, Hart, T.M, Dupuis, A.P, Zamba Campero, M, Nowak, T.A, Stout, J.L, Akopjana, I, Kazaks, A, Bogans, J, Ciota, A.T, Kraiczy, P, Kolokotronis, S.O, Lin, Y.-P. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evolution of an immune evasion determinant shapes pathogen host tropism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6CQB
 
 | |
7QRR
 
 | | Crystal structure of Noumeavirus NMV_189 protein | | Descriptor: | CHLORIDE ION, NMV_189 protein, PHOSPHATE ION | | Authors: | Jeudy, S, Abergel, C. | | Deposit date: | 2022-01-11 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The fibre head structure used by unrelated families of viruses is unexpectedly a major component of the Marseilleviridae and Zamilon virophages capsids
To Be Published
|
|
7ZJM
 
 | | Crystal structure of a complex between CspZ from Borrelia burgdorferi strain B408 and human FH SCR domains 6-7 | | Descriptor: | Complement factor H, CspZ, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Brangulis, K, Marcinkiewicz, A, Hart, T.M, Dupuis, A.P, Zamba Campero, M, Nowak, T.A, Stout, J.L, Akopjana, I, Kazaks, A, Bogans, J, Ciota, A.T, Kraiczy, P, Kolokotronis, S.O, Lin, Y.-P. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural evolution of an immune evasion determinant shapes pathogen host tropism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7LTI
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 2 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-19 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QVN
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with(1H-pyrazol-5-yl)methanol | | Descriptor: | 1~{H}-pyrazol-5-ylmethanol, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
8SSF
 
 | | Minimal protein-only/RNA-free Ribonuclease P from Hydrogenobacter thermophilus | | Descriptor: | RNA-free ribonuclease P, SULFATE ION | | Authors: | Mendoza, J, Mallik, L, Wilhelm, C.A, Koutmos, M. | | Deposit date: | 2023-05-08 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial RNA-free RNase P: Structural and functional characterization of multiple oligomeric forms of a minimal protein-only ribonuclease P.
J.Biol.Chem., 299, 2023
|
|
7LTV
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 3 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QQ7
 
 | |
