6Q0D
 
 | | CRYSTAL STRUCTURE OF LDHA IN COMPLEX WITH COMPOUND NCGC00384414-01 AT 2.05 A RESOLUTION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-[3-(cyclopentylethynyl)-4-fluorophenyl]-5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, GLYCEROL, ... | | Authors: | Dranow, D.M, Davies, D.R. | | Deposit date: | 2019-08-01 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pyrazole-Based Lactate Dehydrogenase Inhibitors with Optimized Cell Activity and Pharmacokinetic Properties.
J.Med.Chem., 63, 2020
|
|
7BGJ
 
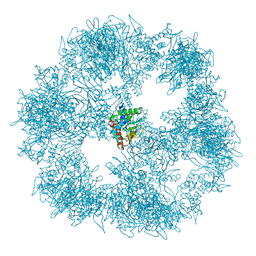 | | C. thermophilum Pyruvate Dehydrogenase Complex Core | | Descriptor: | Acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Tueting, C, Kastritis, P.L. | | Deposit date: | 2021-01-07 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Integrative structure of a 10-megadalton eukaryotic pyruvate dehydrogenase complex from native cell extracts.
Cell Rep, 34, 2021
|
|
6PXW
 
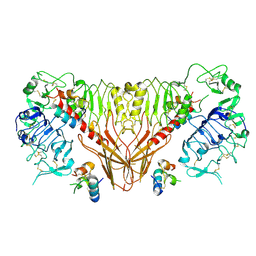 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the top part of the receptor complex. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-28 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
6QXT
 
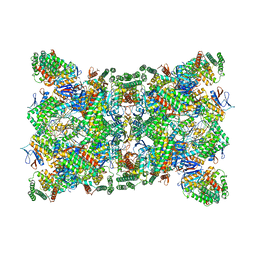 | | Cas1-Cas2-Csn2-DNA dimer complex from the Type II-A CRISPR-Cas system | | Descriptor: | CALCIUM ION, CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, ... | | Authors: | Wilkinson, M, Drabavicius, G, Silanskas, A, Gasiunas, G, Siksnys, V, Wigley, D.B. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the DNA-Bound Spacer Capture Complex of a Type II CRISPR-Cas System.
Mol.Cell, 75, 2019
|
|
6Q2E
 
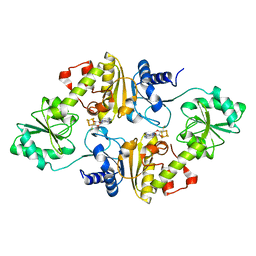 | | Crystal structure of Methanobrevibacter smithii Dph2 bound to 5'-methylthioadenosine | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | The Crystal Structure of Dph2 in Complex with Elongation Factor 2 Reveals the Structural Basis for the First Step of Diphthamide Biosynthesis.
Biochemistry, 58, 2019
|
|
9EQ5
 
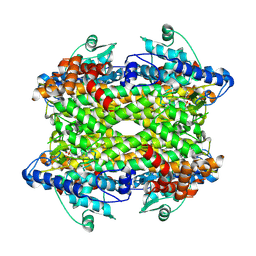 | |
6Q05
 
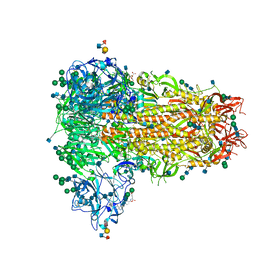 | | MERS-CoV S structure in complex with sialyl-lewisX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
9FAC
 
 | | Additional cryo-EM structure of cardiac amyloid AL59 - mixed polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
7BNT
 
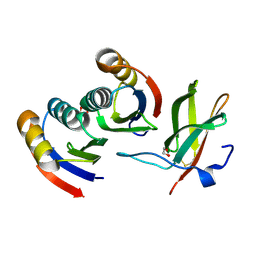 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with a predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Descriptor: | 1,2-ETHANEDIOL, AVR-Pik protein, Predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Authors: | Bialas, A, Langner, T, Harant, A, Contreras, M.P, Stevenson, C.E.M, Lawson, D.M, Sklenar, J, Kellner, R, Moscou, M.J, Terauchi, R, Banfield, M.J, Kamoun, S. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Two NLR immune receptors acquired high-affinity binding to a fungal effector through convergent evolution of their integrated domain.
Elife, 10, 2021
|
|
9CTH
 
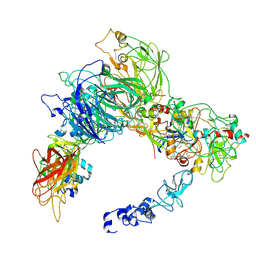 | | Preliminary map of the Prothrombin-prothrombinase complex on nano discs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activated Factor V (FVa) heavy chain, Activated Factor V (FVa) light chain, ... | | Authors: | Stojanovski, B.M, Mohammed, B.M, Di Cera, E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | The Prothrombin-Prothrombinase Interaction.
Subcell Biochem, 104, 2024
|
|
9GAW
 
 | | High-resolution structure of the Anaphase-promoting complex/cyclosome (APC/C) bound to co-activator Cdh1 | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2024-07-29 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structure of the Anaphase-promoting complex (APC/C) bound to co-activator Cdh1
To Be Published
|
|
6Q3Y
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the inhibitor 16i | | Descriptor: | (7~{R})-2-[[2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl]amino]-7-ethyl-5-methyl-8-(phenylmethyl)-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Heidenreich, D, Watts, E, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S, Hoelder, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity.
J.Med.Chem., 62, 2019
|
|
9G2U
 
 | |
9CVG
 
 | |
6QY3
 
 | | Segment of the Cas1-Cas2-Csn2-DNA filament complex from the Type II-A CRISPR-Cas system | | Descriptor: | CALCIUM ION, CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, ... | | Authors: | Wilkinson, M, Drabavicius, G, Silanskas, A, Gasiunas, G, Siksnys, V, Wigley, D.B. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Structure of the DNA-Bound Spacer Capture Complex of a Type II CRISPR-Cas System.
Mol.Cell, 75, 2019
|
|
6Q69
 
 | |
6QCF
 
 | | Ovine respiratory complex I FRC open class 6 | | Descriptor: | Acyl carrier protein, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
9CV9
 
 | | Bufavirus 1 at pH 4.0 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-28 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
7BC3
 
 | | Native virion of Kashmir bee virus at acidic pH | | Descriptor: | Structural polyprotein | | Authors: | Mukhamedova, L, Plevka, P, Fuzik, T, Hrebik, D, Novacek, J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Virion structure and in vitro genome release mechanism of dicistrovirus Kashmir bee virus.
J.Virol., 95, 2021
|
|
9CV0
 
 | | Bufavirus 1 at pH 7.4 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-27 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
9GBV
 
 | |
6QFB
 
 | |
6QGX
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody F7 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoF7, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
9IUY
 
 | |
9FEH
 
 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the STM957 inhibitor | | Descriptor: | Transcription factor ETV6,Guanine-N7 methyltransferase nsp14, ZINC ION, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-pyridin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-cyano-~{N}-ethyl-4-methoxy-benzenesulfonamide | | Authors: | Zilecka, E, Klima, M, Boura, E. | | Deposit date: | 2024-05-20 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of SARS-CoV-2 MTase nsp14 with the inhibitor STM957 reveals inhibition mechanism that is shared with a poxviral MTase VP39.
J Struct Biol X, 10, 2024
|
|
