1TSX
 
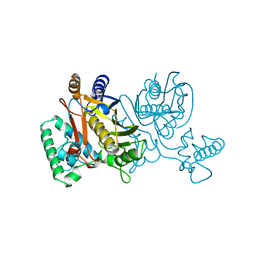 | | THYMIDYLATE SYNTHASE R179E MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
1TSY
 
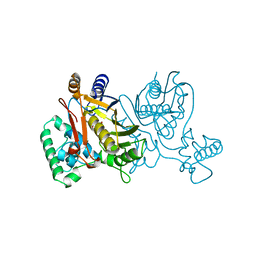 | | THYMIDYLATE SYNTHASE R179K MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
1TSZ
 
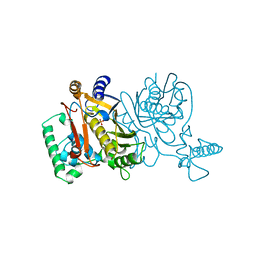 | | THYMIDYLATE SYNTHASE R179K MUTANT | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
1TT0
 
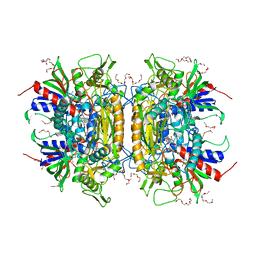 | | Crystal Structure of Pyranose 2-Oxidase | | Descriptor: | ACETATE ION, DODECAETHYLENE GLYCOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hallberg, B.M, Leitner, C, Haltrich, D, Divne, C. | | Deposit date: | 2004-06-21 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the 270 kDa homotetrameric lignin-degrading enzyme pyranose 2-oxidase
J.Mol.Biol., 341, 2004
|
|
1TT1
 
 | |
1TT2
 
 | | Cryogenic crystal structure of Staphylococcal nuclease variant truncated Delta+PHS I92K | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Nguyen, D.M, Reynald, R.L, Gittis, A.G, Lattman, E.E. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and thermodynamic studies of staphylococcal nuclease variants I92E and I92K: insights into polarity of the protein interior
J.Mol.Biol., 341, 2004
|
|
1TT3
 
 | | NMR soulution structure of omega-conotoxin [K10]MVIIA | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-06 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1TT4
 
 | | Structure of NP459575, a predicted glutathione synthase from Salmonella typhimurium | | Descriptor: | MAGNESIUM ION, SULFATE ION, putative cytoplasmic protein | | Authors: | Miller, D.J, Shuvalova, L, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-21 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structure of NP459575, a predicted glutathione synthase from Salmonella typhimurium
To be Published
|
|
1TT5
 
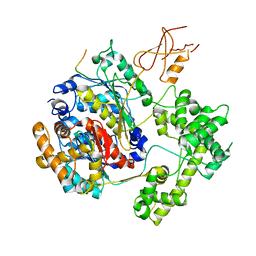 | | Structure of APPBP1-UBA3-Ubc12N26: a unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8 | | Descriptor: | Ubiquitin-conjugating enzyme E2 M, ZINC ION, amyloid protein-binding protein 1, ... | | Authors: | Huang, D.T, Miller, D.W, Mathew, R, Cassell, R, Holton, J.M, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2004-06-21 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1TT6
 
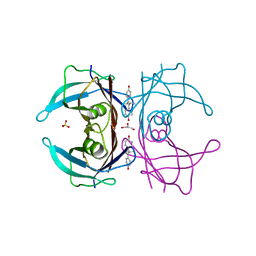 | | The orthorhombic crystal structure of transthyretin in complex with diethylstilbestrol | | Descriptor: | DIETHYLSTILBESTROL, GLYCEROL, SULFATE ION, ... | | Authors: | Morais-de-Sa, E.M, Pereira, P.J.B, Saraiva, M.J, Damas, A.M. | | Deposit date: | 2004-06-22 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of transthyretin in complex with diethylstilbestrol: a promising template for the design of amyloid inhibitors
J.Biol.Chem., 279, 2004
|
|
1TT7
 
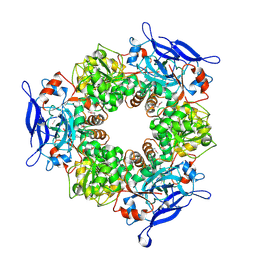 | |
1TT8
 
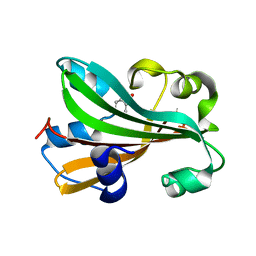 | | CHORISMATE LYASE WITH PRODUCT, 1.0 A RESOLUTION | | Descriptor: | Chorismate-pyruvate lyase, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Vilker, V, Howard, A. | | Deposit date: | 2004-06-22 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural analysis of ligand binding and catalysis in chorismate lyase.
Arch.Biochem.Biophys., 445, 2006
|
|
1TT9
 
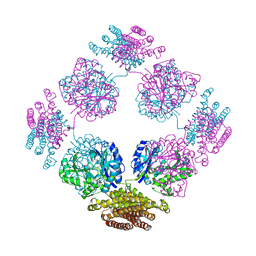 | | Structure of the bifunctional and Golgi associated formiminotransferase cyclodeaminase octamer | | Descriptor: | Formimidoyltransferase-cyclodeaminase (Formiminotransferase- cyclodeaminase) (FTCD) (58 kDa microtubule-binding protein) | | Authors: | Mao, Y, Vyas, N.K, Vyas, M.N, Chen, D.H, Ludtke, S.J, Chiu, W, Quiocho, F.A. | | Deposit date: | 2004-06-22 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structure of the bifunctional and Golgi-associated formiminotransferase cyclodeaminase octamer
Embo J., 23, 2004
|
|
1TTA
 
 | |
1TTB
 
 | |
1TTC
 
 | |
1TTD
 
 | | SOLUTION-STATE STRUCTURE OF A DNA DODECAMER DUPLEX CONTAINING A CIS-SYN THYMINE CYCLOBUTANE DIMER | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*AP*TP*TP*CP*GP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*CP*GP*AP*AP*(TTD)P*AP*AP*G)-3') | | Authors: | Mcateer, K, Jing, Y, Kao, J, Taylor, J.-S, Kennedy, M.A. | | Deposit date: | 1999-01-20 | | Release date: | 1999-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution-state structure of a DNA dodecamer duplex containing a Cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA.
J.Mol.Biol., 282, 1998
|
|
1TTE
 
 | |
1TTF
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE TENTH TYPE III MODULE OF FIBRONECTIN: AN INSIGHT INTO RGD-MEDIATED INTERACTIONS | | Descriptor: | FIBRONECTIN | | Authors: | Main, A.L, Harvey, T.S, Baron, M, Campbell, I.D. | | Deposit date: | 1993-07-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions.
Cell(Cambridge,Mass.), 71, 1992
|
|
1TTG
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE TENTH TYPE III MODULE OF FIBRONECTIN: AN INSIGHT INTO RGD-MEDIATED INTERACTIONS | | Descriptor: | FIBRONECTIN | | Authors: | Main, A.L, Harvey, T.S, Baron, M, Campbell, I.D. | | Deposit date: | 1993-07-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions.
Cell(Cambridge,Mass.), 71, 1992
|
|
1TTH
 
 | | Aspartate Transcarbamoylase Catalytic Chain Mutant Glu50Ala Complexed with N-(Phosphonacetyl-L-Aspartate) (PALA) | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, ... | | Authors: | Stieglitz, K, Stec, B, Baker, D.P, Kantrowitz, E.R. | | Deposit date: | 2004-06-22 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monitoring the Transition from the T to the R State in E.coli Aspartate Transcarbamoylase by X-ray Crystallography: Crystal Structures of the E50A Mutant Enzyme in Four Distinct Allosteric States.
J.Mol.Biol., 341, 2004
|
|
1TTI
 
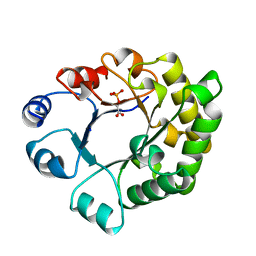 | |
1TTJ
 
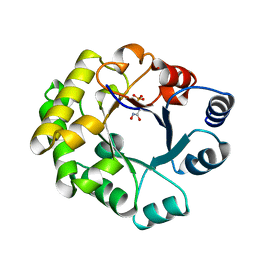 | |
1TTK
 
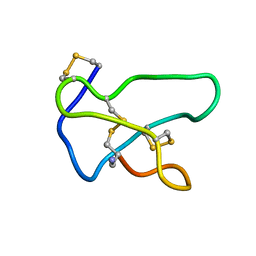 | | NMR solution structure of omega-conotoxin MVIIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-22 | | Release date: | 2004-07-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1TTL
 
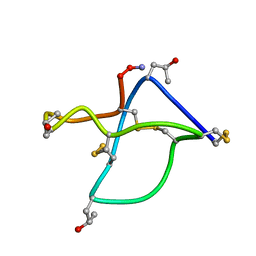 | | Omega-conotoxin GVIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-23 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
