2VV7
 
 | | BJFIXLH IN UNLIGANDED FERROUS FORM | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL, ... | | Authors: | Ayers, R.A, Moffat, K. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Changes in Quaternary Structure in the Signaling Mechanisms of Pas Domains.
Biochemistry, 47, 2008
|
|
5EP3
 
 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain Bound to CV-133 Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,2-dimethylpropyl 2-[(3-oxidanylidene-5-sulfanylidene-2~{H}-1,2,4-triazin-6-yl)amino]ethanoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
5EPL
 
 | | Crystal Structure of chromodomain of CBX4 in complex with inhibitor UNC3866 | | Descriptor: | E3 SUMO-protein ligase CBX4, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
3KLN
 
 | | Vibrio cholerae VpsT | | Descriptor: | Transcriptional regulator, LuxR family | | Authors: | Krasteva, P.V, Navarro, V.A.S, Sondermann, H. | | Deposit date: | 2009-11-08 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.078 Å) | | Cite: | Vibrio cholerae VpsT Regulates Matrix Production and Motility by Directly Sensing Cyclic di-GMP.
Science, 327, 2010
|
|
2WA3
 
 | | FACTOR INHIBITING HIF-1 ALPHA WITH 2-(3-hydroxyphenyl)-2-oxoacetic acid | | Descriptor: | 2-(3-HYDROXYPHENYL)-2-OXO-ETHANOIC ACID, FE (II) ION, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR | | Authors: | Conejo-Garcia, A, Lienard, B.M.R, Clifton, I.J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2009-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for binding of cyclic 2-oxoglutarate analogues to factor-inhibiting hypoxia-inducible factor.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
2WE0
 
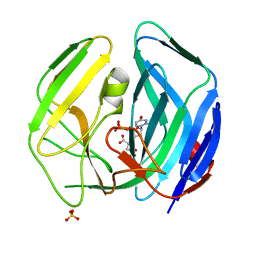 | | EBV dUTPase mutant Cys4Ser | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, MALATE LIKE INTERMEDIATE, ... | | Authors: | Freeman, L, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Flexible Motif V of Epstein-Barr Virus Deoxyuridine 5'-Triphosphate Pyrophosphatase is Essential for Catalysis.
J.Biol.Chem., 284, 2009
|
|
2VYT
 
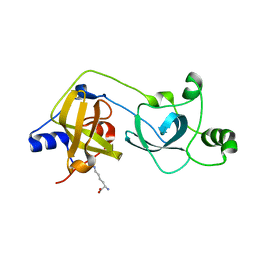 | | The MBT repeats of human SCML2 bind to peptides containing mono methylated lysine. | | Descriptor: | N-METHYL-LYSINE, SEX COMB ON MIDLEG-LIKE PROTEIN 2, TRIETHYLENE GLYCOL | | Authors: | Santiveri, C.M, Lechtenberg, B.C, Allen, M.D, Sathyamurthy, A, Jaulent, A.M, Freund, S.M.V, Bycroft, M. | | Deposit date: | 2008-07-28 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Malignant Brain Tumor Repeats of Human Scml2 Bind to Peptides Containing Monomethylated Lysine.
J.Mol.Biol., 382, 2008
|
|
2W5Y
 
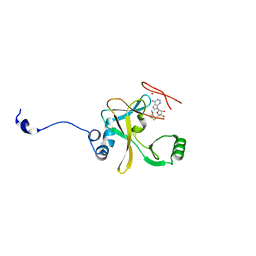 | | Binary Complex of the Mixed Lineage Leukaemia (MLL1) SET Domain with the cofactor product S-Adenosylhomocysteine. | | Descriptor: | HISTONE-LYSINE N-METHYLTRANSFERASE HRX, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Southall, S.M, Wong, P.S, Odho, Z, Roe, S.M, WIlson, J.R. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Requirement of Additional Factors for Mll1 Set Domain Activity and Recognition of Epigenetic Marks.
Mol.Cell, 33, 2009
|
|
7UBJ
 
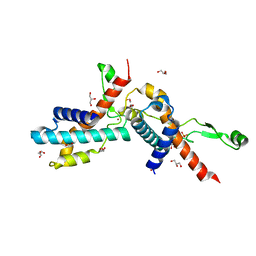 | | Transcription antitermination factor Qlambda, type-I crystal | | Descriptor: | 1,2-ETHANEDIOL, Antitermination protein Q, CHLORIDE ION, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | In transcription antitermination by Q lambda , NusA induces refolding of Q lambda to form a nozzle that extends the RNA polymerase RNA-exit channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UBK
 
 | | Transcription antitermination factor Qlambda, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, Antitermination protein Q, CHLORIDE ION, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | In transcription antitermination by Q lambda , NusA induces refolding of Q lambda to form a nozzle that extends the RNA polymerase RNA-exit channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4YHR
 
 | | Crystal Structure of Yeast Proliferating Cell Nuclear Antigen | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Litman, J.M, Nguyen, V.Q, Kondratick, C.M, Powers, K.T, Schnieders, M.J, Washington, M.T. | | Deposit date: | 2015-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9502 Å) | | Cite: | Dead-End Elimination with a Polarizable Force Field Repacks PCNA Models from Low-Resolution X-ray Diffraction into Atomic Resolution Structures
To be published
|
|
3K1P
 
 | |
6WNH
 
 | | Menin bound to inhibitor M-808 | | Descriptor: | Menin, methyl [(1S,2R)-2-{(1S)-2-(azetidin-1-yl)-1-(3-fluorophenyl)-1-[1-({1-[4-({1-[4-(piperidin-1-yl)butanoyl]azetidin-3-yl}sulfonyl)phenyl]azetidin-3-yl}methyl)piperidin-4-yl]ethyl}cyclopentyl]carbamate, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of M-808 as a Highly Potent, Covalent, Small-Molecule Inhibitor of the Menin-MLL Interaction with StrongIn VivoAntitumor Activity.
J.Med.Chem., 63, 2020
|
|
8BPB
 
 | | Cryo-EM structure of the human SIN3B histone deacetylase core complex at 2.8 Angstrom | | Descriptor: | ACETATE ION, CALCIUM ION, Histone deacetylase 2, ... | | Authors: | Wan, M.S.M, Muhammad, R, Koliopolous, M.G, Alfieri, C. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of assembly, activation and lysine selection by the SIN3B histone deacetylase complex.
Nat Commun, 14, 2023
|
|
8BPC
 
 | | Cryo-EM structure of the human SIN3B histone deacetylase core complex with SAHA at 2.8 Angstrom | | Descriptor: | CALCIUM ION, Histone deacetylase 2, Isoform 2 of Paired amphipathic helix protein Sin3b, ... | | Authors: | Wan, M.S.M, Muhammad, R, Koliopolous, M.G, Alfieri, C. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of assembly, activation and lysine selection by the SIN3B histone deacetylase complex.
Nat Commun, 14, 2023
|
|
8G7Q
 
 | |
8F36
 
 | |
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8G7P
 
 | |
8F37
 
 | |
8GCR
 
 | | HPV16 E6-E6AP-p53 complex | | Descriptor: | Cellular tumor antigen p53, Maltose/maltodextrin-binding periplasmic protein,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Bratkowski, M.A, Wang, J.C.K, Hao, Q, Nile, A.H. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure of the p53 degradation complex from HPV16.
Nat Commun, 15, 2024
|
|
3K1M
 
 | | Crystal Structure of full-length BenM, R156H mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, HTH-type transcriptional regulator benM, ... | | Authors: | Ruangprasert, A, Momany, C, Neidle, E.L, Craven, S.H. | | Deposit date: | 2009-09-28 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of Full-length BenM
To be Published
|
|
3BQY
 
 | | Crystal structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2). | | Descriptor: | ACETIC ACID, PHOSPHATE ION, Putative TetR family transcriptional regulator | | Authors: | Cuff, M.E, Skarina, T, Kagan, O, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-20 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2).
TO BE PUBLISHED
|
|
3K1N
 
 | |
8XP5
 
 | | The Crystal Structure of p53/BCL-xL fusion complex from Biortus. | | Descriptor: | Bcl-2-like protein 1,Cellular tumor antigen p53, ZINC ION | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Bao, C. | | Deposit date: | 2024-01-03 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Crystal Structure of p53/BCL-xL fusion complex from Biortus.
To Be Published
|
|
