7QZ7
 
 | |
7QZ9
 
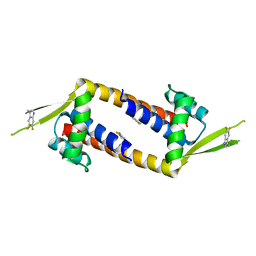 | |
7QZ5
 
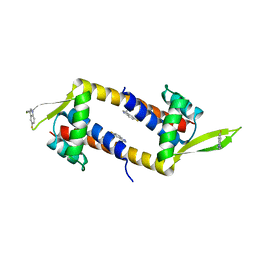 | |
2BNZ
 
 | | Structural basis for cooperative binding of Ribbon-Helix-Helix Omega repressor to inverted DNA heptad repeats | | Descriptor: | 5'-D(*CP*TP*AP*AP*TP*CP*AP*CP*TP*TP *GP*TP*GP*AP*TP*TP*CP*G)-3', 5'-D(*GP*AP*AP*TP*CP*AP*CP*AP*AP*GP *TP*GP*AP*TP*TP*AP*GP*C)-3', ORF OMEGA | | Authors: | Weihofen, W.A, Cicek, A, Pratto, F, Alonso, J.C, Saenger, W. | | Deposit date: | 2005-04-06 | | Release date: | 2006-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of Omega Repressors Bound to Direct and Inverted DNA Repeats Explain Modulation of Transcription.
Nucleic Acids Res., 34, 2006
|
|
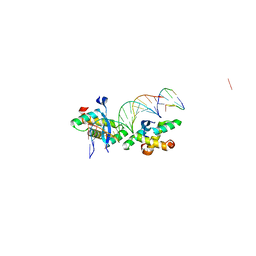
2BNW
 
 | | Structural basis for cooperative binding of Ribbon-Helix-Helix Omega repressor to direct DNA heptad repeats | | Descriptor: | 5'-D(*CP*TP*TP*GP*TP*GP*AP*TP*TP*TP *GP*TP*GP*AP*TP*TP*CP*G)-3', 5'-D(*GP*AP*AP*TP*CP*AP*CP*AP*AP*AP *TP*CP*AP*CP*AP*AP*GP*C)-3', ORF OMEGA | | Authors: | Weihofen, W.A, Cicek, A, Pratto, F, Alonso, J.C, Saenger, W. | | Deposit date: | 2005-04-05 | | Release date: | 2006-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of Omega Repressors Bound to Direct and Inverted DNA Repeats Explain Modulation of Transcription.
Nucleic Acids Res., 34, 2006
|
|
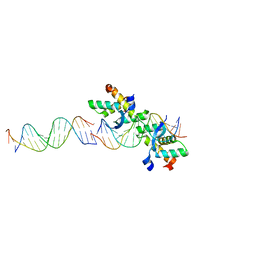
3P9T
 
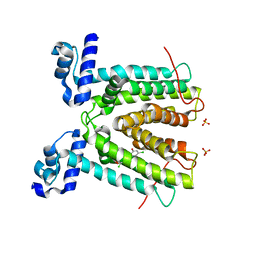 | | SmeT-Triclosan complex | | Descriptor: | Repressor, SULFATE ION, TRICLOSAN | | Authors: | Hernandez, A, Ruiz, F.M, Romero, A, Martinez, J.L. | | Deposit date: | 2010-10-18 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Binding of Triclosan to SmeT, the Repressor of the Multidrug Efflux Pump SmeDEF, Induces Antibiotic Resistance in Stenotrophomonas maltophilia.
Plos Pathog., 7, 2011
|
|
7F5W
 
 | |
1YPV
 
 | |
3EHI
 
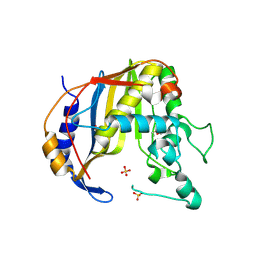 | | Crystal Structure of Human Thymidyalte Synthase M190K with Loop 181-197 stabilized in the inactive conformation | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Thymidylate synthase | | Authors: | Lovelace, L.L, Gibson, L.M, Johnson, S.R, Berger, S.H, Lebioda, L. | | Deposit date: | 2008-09-12 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Variants of human thymidylate synthase with loop 181-197 stabilized in the inactive conformation.
Protein Sci., 18, 2009
|
|
2ONB
 
 | | Human Thymidylate Synthase at low salt conditions with PDPA bound | | Descriptor: | 1,2-ETHANEDIOL, PROPANE-1,3-DIYLBIS(PHOSPHONIC ACID), SULFATE ION, ... | | Authors: | Lovelace, L.L, Gibson, L.M, Lebioda, L. | | Deposit date: | 2007-01-23 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative Inhibition of Human Thymidylate Synthase by Mixtures of Active Site Binding and Allosteric Inhibitors
Biochemistry, 46, 2007
|
|
3WIO
 
 | | Crystal structure of OSD14 in complex with hydroxy D-ring | | Descriptor: | (5R)-5-hydroxy-3-methylfuran-2(5H)-one, Probable strigolactone esterase D14 | | Authors: | Xue, Y.-L, Miyakawa, T, Hou, F, Qin, H.-M, Tanokura, M. | | Deposit date: | 2013-09-22 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of strigolactone perception by DWARF14
Nat Commun, 4, 2013
|
|
6CBQ
 
 | | Crystal structure of QscR bound to agonist S3 | | Descriptor: | (2S)-2-hexyl-N-[(3S)-2-oxooxolan-3-yl]decanamide, LuxR family transcriptional regulator | | Authors: | Churchill, M.E.A, Wysoczynski-Horita, C.L. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of agonism and antagonism of the Pseudomonas aeruginosa quorum sensing regulator QscR with non-native ligands.
Mol. Microbiol., 108, 2018
|
|
8A39
 
 | | Crystal Structure of PaaX from Escherichia coli W | | Descriptor: | DNA-binding transcriptional repressor of phenylacetic acid degradation, aryl-CoA responsive, GLYCEROL, ... | | Authors: | Molina, R, Alba-Perez, A, Hermoso, J.A. | | Deposit date: | 2022-06-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of PaaX, the main repressor of the phenylacetate degradation pathway in Escherichia coli W: A novel fold of transcription regulator proteins.
Int.J.Biol.Macromol., 254, 2024
|
|
5D4S
 
 | | Crystal Structure of AraR(DBD) in complex with operator ORX1 | | Descriptor: | Arabinose metabolism transcriptional repressor, DNA (5'-D(*AP*AP*AP*TP*AP*CP*AP*TP*AP*CP*GP*TP*AP*CP*AP*AP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*AP*TP*TP*TP*GP*TP*AP*CP*GP*TP*AP*TP*GP*TP*AP*TP*T)-3') | | Authors: | Jain, D, Narayanan, N, Nair, D.T. | | Deposit date: | 2015-08-08 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Plasticity in Repressor-DNA Interactions Neutralizes Loss of Symmetry in Bipartite Operators.
J.Biol.Chem., 291, 2016
|
|
5D4R
 
 | | Crystal Structure of AraR(DBD) in complex with operator ORE1 | | Descriptor: | Arabinose metabolism transcriptional repressor, DNA (5'-D(*AP*TP*AP*TP*TP*TP*GP*TP*AP*CP*GP*TP*AP*CP*TP*AP*AP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*TP*AP*GP*TP*AP*CP*GP*TP*AP*CP*AP*AP*AP*TP*A)-3') | | Authors: | Jain, D, Narayanan, N, Nair, D.T. | | Deposit date: | 2015-08-08 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Plasticity in Repressor-DNA Interactions Neutralizes Loss of Symmetry in Bipartite Operators.
J.Biol.Chem., 291, 2016
|
|
8QA9
 
 | | Crystal structure of the RK2 plasmid encoded co-complex of the C-terminally truncated transcriptional repressor protein KorB complexed with the partner repressor protein KorA bound to OA-DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*TP*TP*AP*GP*CP*TP*AP*AP*AP*CP*A)-3'), SULFATE ION, Transcriptional repressor protein KorB, ... | | Authors: | McLean, T.C, Mundy, J.E.A, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2023-08-22 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the RK2 plasmid encoded co-complex of the C-terminally truncated transcriptional repressor protein KorB complexed with the partner repressor protein KorA bound to OA-DNA
To Be Published
|
|
6J05
 
 | | Structures of two ArsR As(III)-responsive repressors: implications for the mechanism of derepression | | Descriptor: | ARSENIC, SODIUM ION, Transcriptional regulator ArsR | | Authors: | Prabaharan, C, Kandavelu, P, Packianathan, C, Rosen, P.B, Thiyagarajan, S. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of two ArsR As(III)-responsive transcriptional repressors: Implications for the mechanism of derepression.
J.Struct.Biol., 207, 2019
|
|
7DOB
 
 | |
7DOA
 
 | |
4U52
 
 | | Crystal structure of Nagilactone C bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
6WMQ
 
 | |
8QA8
 
 | |
6FPL
 
 | | TETR(D) E147A MUTANT IN COMPLEX WITH TETRACYCLINE AND MAGNESIUM | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, TETRACYCLINE, ... | | Authors: | Hinrichs, W, Palm, G.J, Berndt, L, Girbardt, B. | | Deposit date: | 2018-02-11 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
6FPM
 
 | | TetR(D) T103A mutant in complex with tetracycline and magnesium | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, TETRACYCLINE, ... | | Authors: | Hinrichs, W, Palm, G.J, Berndt, L, Girbardt, B. | | Deposit date: | 2018-02-11 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
6FTS
 
 | | TETR(D) N82A MUTANT IN COMPLEX WITH PEG4 | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, Tetracycline repressor protein class D | | Authors: | Hinrichs, W, Palm, G.J, Berndt, L, Girbardt, B. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
