4LT2
 
 | | HEWL co-crystallized with Carboplatin in non-NaCl conditions: crystal 2 processed using the EVAL software package | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carboplatin binding to a model protein in non-NaCl conditions to eliminate partial conversion to cisplatin, and the use of different criteria to choose the resolution limit
To be Published
|
|
1T6G
 
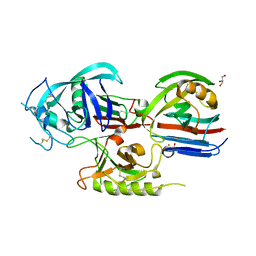 | | Crystal structure of the Triticum aestivum xylanase inhibitor-I in complex with aspergillus niger xylanase-I | | Descriptor: | Endo-1,4-beta-xylanase I, GLYCEROL, xylanase inhibitor | | Authors: | Sansen, S, De Ranter, C.J, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2004-05-06 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for inhibition of Aspergillus niger xylanase by triticum aestivum xylanase inhibitor-I
J.Biol.Chem., 279, 2004
|
|
1TLS
 
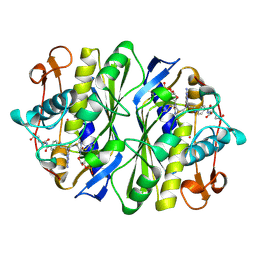 | | THYMIDYLATE SYNTHASE TERNARY COMPLEX WITH FDUMP AND METHYLENETETRAHYDROFOLATE | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Hyatt, D.C, Maley, F, Montfort, W.R. | | Deposit date: | 1996-12-03 | | Release date: | 1997-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Use of strain in a stereospecific catalytic mechanism: crystal structures of Escherichia coli thymidylate synthase bound to FdUMP and methylenetetrahydrofolate.
Biochemistry, 36, 1997
|
|
3NHW
 
 | |
7D4R
 
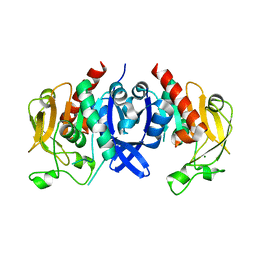 | | SpuA native structure | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1T0Q
 
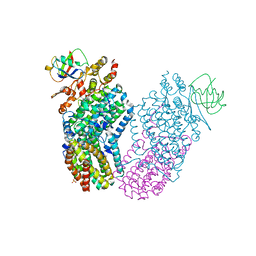 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase | | Descriptor: | FE (III) ION, HYDROXIDE ION, SULFANYLACETIC ACID, ... | | Authors: | Sazinsky, M.H, Bard, J, Di Donato, A, Lippard, S.J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase from Pseudomonas stutzeri OX1: INSIGHT INTO THE SUBSTRATE SPECIFICITY, SUBSTRATE CHANNELING, AND ACTIVE SITE TUNING OF MULTICOMPONENT MONOOXYGENASES.
J.Biol.Chem., 279, 2004
|
|
3NKE
 
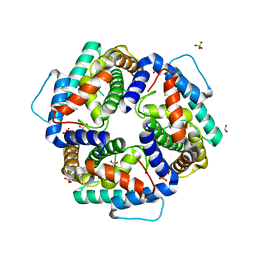 | | High resolution structure of the C-terminal domain CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, SULFITE ION, ... | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
5IJZ
 
 | |
1T8F
 
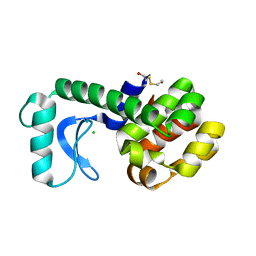 | | Crystal structure of phage T4 lysozyme mutant R14A/K16A/I17A/K19A/T21A/E22A/C54T/C97A | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | He, M.M, Wood, Z.A, Baase, W.A, Xiao, H, Matthews, B.W. | | Deposit date: | 2004-05-12 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation.
Protein Sci., 13, 2004
|
|
1T26
 
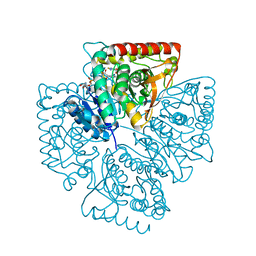 | | Plasmodium falciparum lactate dehydrogenase complexed with NADH and 4-hydroxy-1,2,5-thiadiazole-3-carboxylic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-HYDROXY-1,2,5-THIADIAZOLE-3-CARBOXYLIC ACID, L-lactate dehydrogenase | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
7D50
 
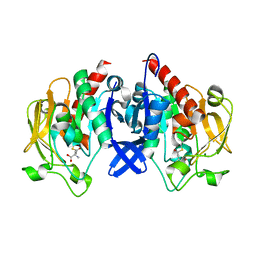 | | SpuA mutant - H221N with glutamyl-thioester | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ILF
 
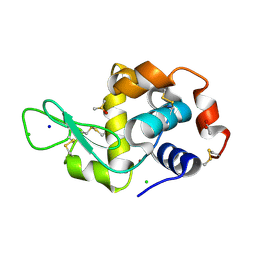 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme and compound 4, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2016-03-04 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
1T9G
 
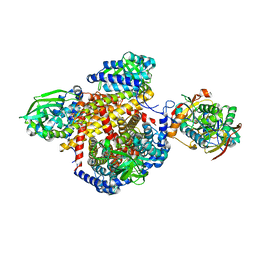 | | Structure of the human MCAD:ETF complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Toogood, H.S, van Thiel, A, Basran, J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2004-05-17 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Extensive domain motion and electron transfer in the human electron transferring flavoprotein-medium chain Acyl-CoA dehydrogenase complex
J.Biol.Chem., 279, 2004
|
|
7D53
 
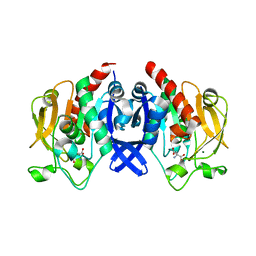 | | SpuA mutant - H221N with Glu | | Descriptor: | GLUTAMIC ACID, MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1T5F
 
 | | arginase I-AOH complex | | Descriptor: | (S)-2-AMINO-7,7-DIHYDROXYHEPTANOIC ACID, Arginase 1, MANGANESE (II) ION | | Authors: | Shin, H, Cama, E, Christianson, D.W. | | Deposit date: | 2004-05-04 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of amino acid aldehydes as transition-state analogue inhibitors of arginase
J.Am.Chem.Soc., 126, 2004
|
|
5I81
 
 | | aSMase with zinc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Zhou, Y.F, Wei, R.R. | | Deposit date: | 2016-02-18 | | Release date: | 2016-09-07 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Human acid sphingomyelinase structures provide insight to molecular basis of Niemann-Pick disease.
Nat Commun, 7, 2016
|
|
5I8Q
 
 | | S. cerevisiae Prp43 in complex with RNA and ADPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP43, ... | | Authors: | He, Y, Nielsen, K.H, Andersen, G.R. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of the DEAH/RHA ATPase Prp43p bound to RNA implicates a pair of hairpins and motif Va in translocation along RNA.
RNA, 23, 2017
|
|
4M56
 
 | | The Structure of Wild-type MalL from Bacillus subtilis | | Descriptor: | D-glucose, GLYCEROL, Oligo-1,6-glucosidase 1, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4XFA
 
 | |
3N9A
 
 | |
4XFI
 
 | |
3N9C
 
 | | Mite-y Lysozyme: Marmite | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mite-y Lysozyme Crystals and Structures
To be Published
|
|
5IP5
 
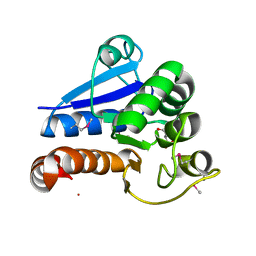 | |
3N9K
 
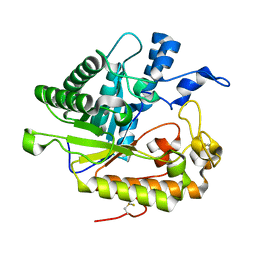 | | F229A/E292S Double Mutant of Exo-beta-1,3-glucanase from Candida albicans in Complex with Laminaritriose at 1.7 A | | Descriptor: | CALCIUM ION, Glucan 1,3-beta-glucosidase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2010-05-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carbohydrate binding sites in Candida albicans exo-beta-1,3-glucanase and the role of the Phe-Phe 'clamp' at the active site entrance
Febs J., 277, 2010
|
|
1FAE
 
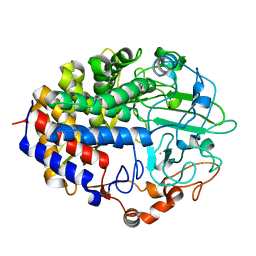 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellobiose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-13 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
