1T3U
 
 | | Unknown conserved bacterial protein from Pseudomonas aeruginosa PAO1 | | Descriptor: | conserved hypothetical protein | | Authors: | Rajashankar, K.R, Kneiwel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-04 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a conserved hypothetical protein Pseudomonas aeruginosa PA01
To be Published
|
|
1T3V
 
 | | The NMR solution structure of TM1816 | | Descriptor: | conserved hypothetical protein | | Authors: | Columbus, L, Peti, W, Herrmann, T, Etazady, T, Klock, H, Lesley, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-04-27 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of the conserved hypothetical protein TM1816 from Thermotoga maritima.
Proteins, 60, 2005
|
|
1T3W
 
 | | Crystal Structure of the E.coli DnaG C-terminal domain (residues 434 to 581) | | Descriptor: | ACETIC ACID, DNA primase | | Authors: | Oakley, A.J, Loscha, K.V, Schaeffer, P.M, Liepinsh, E, Wilce, M.C.J, Otting, G, Dixon, N.E. | | Deposit date: | 2004-04-28 | | Release date: | 2004-11-02 | | Last modified: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal and solution structures of the helicase-binding domain of Escherichia coli primase
J.Biol.Chem., 280, 2005
|
|
1T3X
 
 | |
1T3Y
 
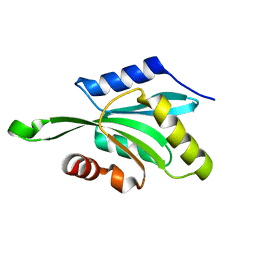 | |
1T3Z
 
 | | Formyl-CoA Tranferase mutant Asp169 to Ser | | Descriptor: | Formyl-coenzyme A transferase, OXIDIZED COENZYME A | | Authors: | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
1T40
 
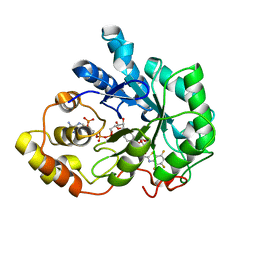 | | Crystal structure of human aldose reductase complexed with NADP and IDD552 at ph 5 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [5-FLUORO-2-({[(4,5,7-TRIFLUORO-1,3-BENZOTHIAZOL-2-YL)METHYL]AMINO}CARBONYL)PHENOXY]ACETIC ACID | | Authors: | Ruiz, F, Hazemann, I, Mitschler, A, Chevrier, B, Schneider, T, Joachimiak, A, Karplus, M, Podjarny, A. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystallographic structure of the aldose reductase-IDD552 complex shows direct proton donation from tyrosine 48.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T41
 
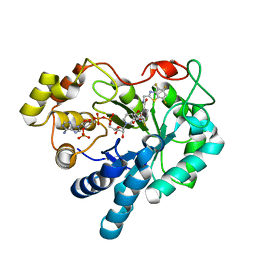 | | Crystal structure of human aldose reductase complexed with NADP and IDD552 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [5-FLUORO-2-({[(4,5,7-TRIFLUORO-1,3-BENZOTHIAZOL-2-YL)METHYL]AMINO}CARBONYL)PHENOXY]ACETIC ACID | | Authors: | Ruiz, F, Hazemann, I, Mitschler, A, Chevrier, B, Schneider, T, Joachimiak, A, Karplus, M, Podjarny, A. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystallographic structure of the aldose reductase-IDD552 complex shows direct proton donation from tyrosine 48.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T43
 
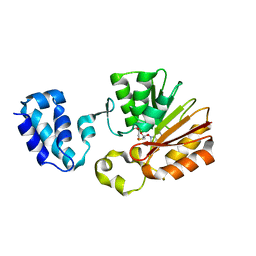 | | Crystal Structure Analysis of E.coli Protein (N5)-Glutamine Methyltransferase (HemK) | | Descriptor: | Protein methyltransferase hemK, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yang, Z, Shipman, L, Zhang, M, Anton, B.P, Roberts, R.J, Cheng, X. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural characterization and comparative phylogenetic analysis of Escherichia coli HemK, a protein (N5)-glutamine methyltransferase.
J.Mol.Biol., 340, 2004
|
|
1T44
 
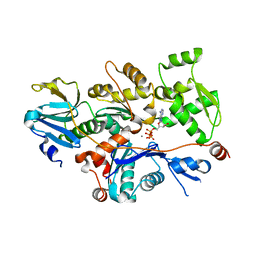 | | Structural basis of actin sequestration by thymosin-B4: Implications for arp2/3 activation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha, ... | | Authors: | Irobi, E, Aguda, A.H, Larsson, M, Burtnick, L.D, Robinson, R.C. | | Deposit date: | 2004-04-28 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of actin sequestration by thymosin-beta4: implications for WH2 proteins.
Embo J., 23, 2004
|
|
1T45
 
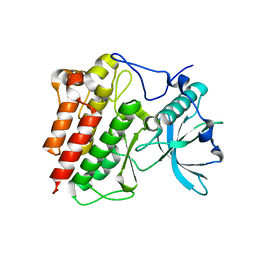 | | STRUCTURAL BASIS FOR THE AUTOINHIBITION AND STI-571 INHIBITION OF C-KIT TYROSINE KINASE | | Descriptor: | Homo sapiens v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | | Authors: | Mol, C.D, Dougan, D.R, Schneider, T.R, Skene, R.J, Kraus, M.L, Scheibe, D.N, Snell, G.P, Zou, H, Sang, B.C, Wilson, K.P. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase.
J.Biol.Chem., 279, 2004
|
|
1T46
 
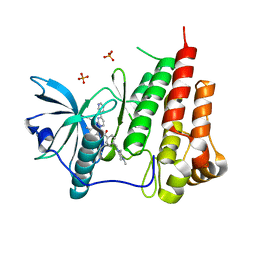 | | STRUCTURAL BASIS FOR THE AUTOINHIBITION AND STI-571 INHIBITION OF C-KIT TYROSINE KINASE | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Homo sapiens v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog, PHOSPHATE ION | | Authors: | Mol, C.D, Dougan, D.R, Schneider, T.R, Skene, R.J, Kraus, M.L, Scheibe, D.N, Snell, G.P, Zou, H, Sang, B.C, Wilson, K.P. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase.
J.Biol.Chem., 279, 2004
|
|
1T47
 
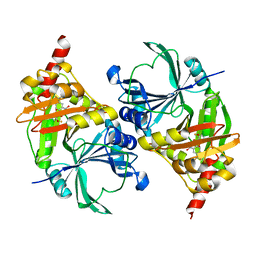 | | Structure of fe2-HPPD bound to NTBC | | Descriptor: | 2-{HYDROXY[2-NITRO-4-(TRIFLUOROMETHYL)PHENYL]METHYLENE}CYCLOHEXANE-1,3-DIONE, 4-hydroxyphenylpyruvate dioxygenase, FE (II) ION | | Authors: | Brownlee, J, Johnson-Winters, K, Harrison, D.H.T, Moran, G.R. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Ferrous Form of (4-Hydroxyphenyl)pyruvate Dioxygenase from Streptomyces avermitilis in Complex with the Therapeutic Herbicide, NTBC
Biochemistry, 43, 2004
|
|
1T48
 
 | | Allosteric Inhibition of Protein Tyrosine Phosphatase 1B | | Descriptor: | 3-(3,5-DIBROMO-4-HYDROXY-BENZOYL)-2-ETHYL-BENZOFURAN-6-SULFONIC ACID DIMETHYLAMIDE, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Wiesmann, C, Barr, K.J, Kung, J, Zhu, J, Shen, W, Fahr, B.J, Zhong, M, Erlanson, D.A, Taylor, L, Randal, M, McDowell, R.S, Hansen, S.K. | | Deposit date: | 2004-04-28 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric inhibition of protein tyrosine phosphatase 1B
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T49
 
 | | Allosteric Inhibition of Protein Tyrosine Phosphatase 1B | | Descriptor: | 3-(3,5-DIBROMO-4-HYDROXY-BENZOYL)-2-ETHYL-BENZOFURAN-6-SULFONIC ACID (4-SULFAMOYL-PHENYL)-AMIDE, MAGNESIUM ION, Protein-tyrosine phosphatase, ... | | Authors: | Wiesmann, C, Barr, K.J, Kung, J, Zhu, J, Shen, W, Fahr, B.J, Zhong, M, Taylor, L, Randal, M, McDowell, R.S, Hansen, S.K. | | Deposit date: | 2004-04-28 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric inhibition of protein tyrosine phosphatase 1B.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T4A
 
 | | Structure of B. Subtilis PurS C2 Crystal Form | | Descriptor: | PurS | | Authors: | Anand, R, Hoskins, A.A, Bennett, E.M, Sintchak, M.D, Stubbe, J, Ealick, S.E. | | Deposit date: | 2004-04-28 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A model for the Bacillus subtilis formylglycinamide ribonucleotide amidotransferase multiprotein complex.
Biochemistry, 43, 2004
|
|
1T4B
 
 | | 1.6 Angstrom structure of Esherichia coli aspartate-semialdehyde dehydrogenase. | | Descriptor: | Aspartate-semialdehyde dehydrogenase, SODIUM ION | | Authors: | Nichols, C.E, Dhaliwal, B, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-04-29 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution Structures Reveal Details of Domain Closure and "Half-of-sites-reactivity" in Escherichia coli Aspartate beta-Semialdehyde Dehydrogenase.
J.Mol.Biol., 341, 2004
|
|
1T4C
 
 | | Formyl-CoA Transferase in complex with Oxalyl-CoA | | Descriptor: | COENZYME A, Formyl-CoA:oxalate CoA-transferase, OXALIC ACID | | Authors: | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | Deposit date: | 2004-04-29 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
1T4D
 
 | | Crystal structure of Escherichia coli aspartate beta-semialdehyde dehydrogenase (EcASADH), at 1.95 Angstrom resolution | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Nichols, C.E, Dhaliwal, B, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-04-29 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution Structures Reveal Details of Domain Closure and "Half-of-sites-reactivity" in Escherichia coli Aspartate beta-Semialdehyde Dehydrogenase.
J.Mol.Biol., 341, 2004
|
|
1T4E
 
 | | Structure of Human MDM2 in complex with a Benzodiazepine Inhibitor | | Descriptor: | (4-CHLOROPHENYL)[3-(4-CHLOROPHENYL)-7-IODO-2,5-DIOXO-1,2,3,5-TETRAHYDRO-4H-1,4-BENZODIAZEPIN-4-YL]ACETIC ACID, Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Grasberger, B.L, Schubert, C, Koblish, H.K, Carver, T.E, Franks, C.F, Zhao, S.Y, Lu, T, LaFrance, L.V, Parks, D.J. | | Deposit date: | 2004-04-29 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells
J.Med.Chem., 48, 2005
|
|
1T4F
 
 | | Structure of human MDM2 in complex with an optimized p53 peptide | | Descriptor: | SULFATE ION, Ubiquitin-protein ligase E3 Mdm2, optimized p53 peptide | | Authors: | Grasberger, B.L, Schubert, C, Koblish, H.K, Carver, T.E, Franks, C.F, Zhao, S.Y, Lu, T, LaFrance, L.V, Parks, D.J. | | Deposit date: | 2004-04-29 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells
J.Med.Chem., 48, 2005
|
|
1T4G
 
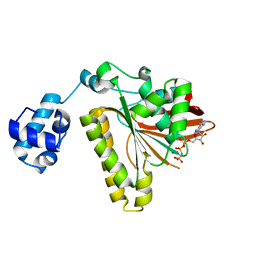 | | ATPase in complex with AMP-PNP | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, Y, He, Y, Moya, I.A, Qian, X, Luo, Y. | | Deposit date: | 2004-04-29 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Archaeal Recombinase RadA; A Snapshot of Its Extended Conformation.
Mol.Cell, 15, 2004
|
|
1T4I
 
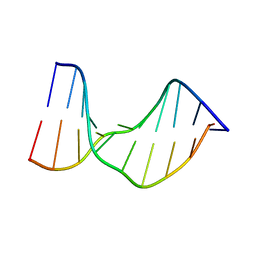 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*AP*TP*TP*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.S, Kang, C. | | Deposit date: | 2004-04-29 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a DNA Decamer Containing a cis-syn Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1T4J
 
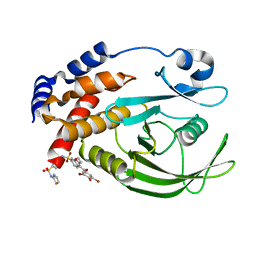 | | Allosteric Inhibition of Protein Tyrosine Phosphatase 1B | | Descriptor: | 3-(3,5-DIBROMO-4-HYDROXY-BENZOYL)-2-ETHYL-BENZOFURAN-6-SULFONIC ACID [4-(THIAZOL-2-YLSULFAMOYL)-PHENYL]-AMIDE, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Wiesmann, C, Barr, K.J, Kung, J, Zhu, J, Shen, W, Fahr, B.J, Zhong, M, Taylor, L, Randal, M, McDowell, R.S, Hansen, S.K. | | Deposit date: | 2004-04-29 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric inhibition of protein tyrosine phosphatase 1B
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T4K
 
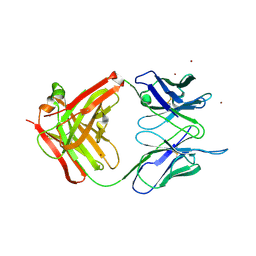 | | Crystal Structure of Unliganded Aldolase Antibody 93F3 Fab | | Descriptor: | IMMUNOGLOBULIN IGG1, HEAVY CHAIN, KAPPA LIGHT CHAIN, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2004-04-29 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Origin of Enantioselectivity in Aldolase Antibodies: Crystal Structure, Site-directed Mutagenesis, and Computational Analysis
J.Mol.Biol., 343, 2004
|
|
