4XWN
 
 | |
4XLZ
 
 | | N,N'-diacetylchitobiose deacetylase (SeMet derivative) from Pyrococcus furiosus in the presence of cadmium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5IDW
 
 | |
7D89
 
 | |
3N5N
 
 | |
5IEF
 
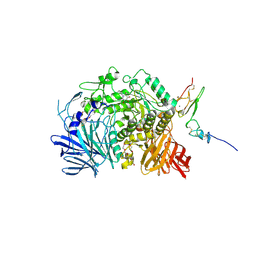 | | Murine endoplasmic reticulum alpha-glucosidase II with N-butyl-1-deoxynojirimycin | | Descriptor: | (2R,3R,4R,5S)-1-BUTYL-2-(HYDROXYMETHYL)PIPERIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5I4X
 
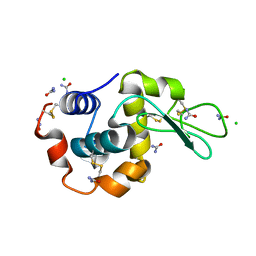 | |
4M29
 
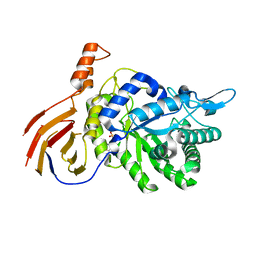 | | Structure of a GH39 Beta-xylosidase from Caulobacter crescentus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-xylosidase | | Authors: | Polo, C.C, Santos, C.R, Correa, J.M, Simao, R.C.G, Seixas, F.A.V, Murakami, M.T. | | Deposit date: | 2013-08-05 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a GH39 Beta-xylosidase from Caulobacter crescentus
Thesis
|
|
3N80
 
 | |
1F95
 
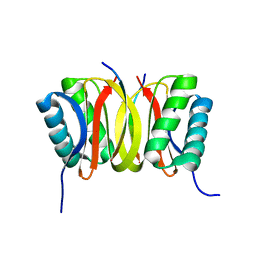 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND BIM PEPTIDE COMPLEX | | Descriptor: | BCL2-LIKE 11 (APOPTOSIS FACILITATOR), DYNEIN | | Authors: | Fan, J.-S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
4X9Y
 
 | |
3NA0
 
 | | Crystal structure of human CYP11A1 in complex with 20,22-dihydroxycholesterol | | Descriptor: | (3alpha,8alpha,22R)-cholest-5-ene-3,20,22-triol, Adrenodoxin, mitochondrial, ... | | Authors: | Strushkevich, N.V, MacKenzie, F, Tempel, W, Botchkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J.U, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2010-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NAR
 
 | | Crystal structure of ZHX1 HD4 (zinc-fingers and homeoboxes protein 1, homeodomain 4) | | Descriptor: | SULFATE ION, Zinc fingers and homeoboxes protein 1 | | Authors: | Ren, J, Bird, L.E, Owens, R.J, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel structural features in two ZHX homeodomains derived from a systematic study of single and multiple domains
Bmc Struct.Biol., 10, 2010
|
|
1FEA
 
 | |
4M1J
 
 | | Crystal structure of Pseudomonas aeruginosa PvdQ in complex with a transition state analogue | | Descriptor: | Acyl-homoserine lactone acylase PvdQ subunit alpha, Acyl-homoserine lactone acylase PvdQ subunit beta, GLYCEROL, ... | | Authors: | Wu, R, Clevenger, K, Er, J, Fast, W.L, Liu, D. | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Design of a Transition State Analogue with Picomolar Affinity for Pseudomonas aeruginosa PvdQ, a Siderophore Biosynthetic Enzyme.
Acs Chem.Biol., 8, 2013
|
|
4XAF
 
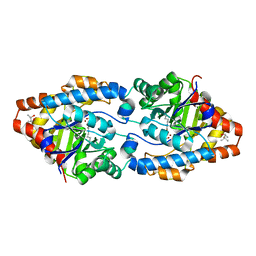 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R1, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-14 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4M24
 
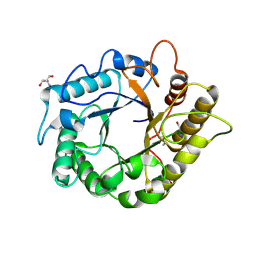 | | Crystal structure of the endo-1,4-glucanase, RBcel1, in complex with cellobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Delsaute, M, Berlemont, R, Van elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-08-05 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Characterisation of two GH family 5 cellulases required for bacterial cellulose production
To be Published
|
|
4M33
 
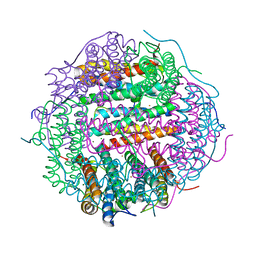 | | Crystal structure of gated-pore mutant H141D of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
5IK0
 
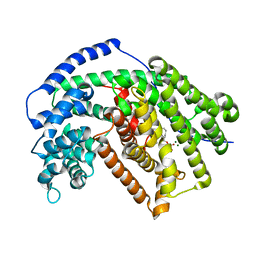 | | Tobacco 5-epi-aristolochene synthase with FPP | | Descriptor: | 5-epi-aristolochene synthase, FARNESYL DIPHOSPHATE, MAGNESIUM ION | | Authors: | Koo, H.J, Xu, Y, Louie, G.V, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthetic potential of sesquiterpene synthases: product profiles of Egyptian Henbane premnaspirodiene synthase and related mutants.
J.Antibiot., 69, 2016
|
|
1F6W
 
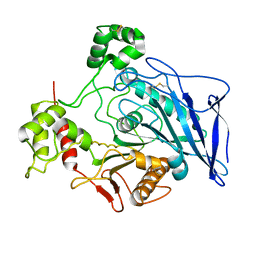 | |
5IK9
 
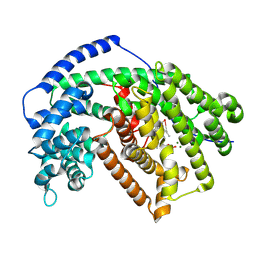 | | Tobacco 5-epi-aristolochene with farnesyl monophosphate | | Descriptor: | (2E,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl dihydrogen phosphate, 5-epi-aristolochene synthase, MAGNESIUM ION | | Authors: | Koo, H.J, Xu, Y, Louie, G.V, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Biosynthetic potential of sesquiterpene synthases: product profiles of Egyptian Henbane premnaspirodiene synthase and related mutants.
J.Antibiot., 69, 2016
|
|
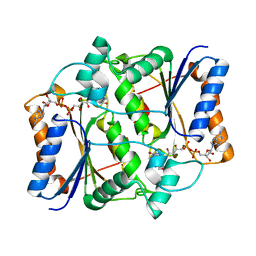
3NHS
 
 | |
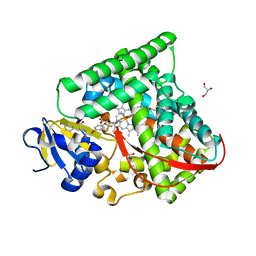
7CZI
 
 | |
4XEE
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-23 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
7D0T
 
 | |
