5SV0
 
 | | Structure of the ExbB/ExbD complex from E. coli at pH 7.0 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Biopolymer transport protein ExbB, CALCIUM ION | | Authors: | Celia, H, Botos, I, Lloubes, R, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2016-08-04 | | Release date: | 2016-09-28 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into the role of the Ton complex in energy transduction.
Nature, 538, 2016
|
|
5SYA
 
 | |
1U4A
 
 | | Solution structure of human SUMO-3 C47S | | Descriptor: | Ubiquitin-like protein SMT3A | | Authors: | Ding, H, Xu, Y, Dai, H, Tang, Y, Wu, J, Shi, Y. | | Deposit date: | 2004-07-23 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human SUMO-3 C47S and Its Binding Surface for Ubc9
Biochemistry, 44, 2005
|
|
1U67
 
 | | Crystal Structure of Arachidonic Acid Bound to a Mutant of Prostagladin H Synthase-1 that Forms Predominantly 11-HPETE. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, PROTOPORPHYRIN IX CONTAINING CO, ... | | Authors: | Harman, C.A, Rieke, C.J, Garavito, R.M, Smith, W.L. | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of arachidonic Acid bound to a mutant of prostaglandin endoperoxide h synthase-1 that forms predominantly 11-hydroperoxyeicosatetraenoic Acid.
J.Biol.Chem., 279, 2004
|
|
5T0U
 
 | | CTCF ZnF2-7 and DNA complex structure | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Hashimoto, H, Wang, D, Cheng, X. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
1TNZ
 
 | | Rat Protein Geranylgeranyltransferase Type-I Complexed with a GGPP analog and a RRCVLL Peptide Derived from Cdc42 splice isoform-2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[METHYL-(5-GERANYL-4-METHYL-PENT-3-ENYL)-AMINO]-ETHYL-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Reid, T.S, Terry, K.L, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity.
J.Mol.Biol., 343, 2004
|
|
1U7L
 
 | |
5T4B
 
 | | Human DPP4 in complex with a ligand 34a | | Descriptor: | 2-[(3R)-3-aminopiperidin-1-yl]-3-(but-2-yn-1-yl)-5-[(4-methylquinazolin-2-yl)methyl]-3H-imidazo[2,1-b]purin-4(5H)-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Scaffold-hopping from xanthines to tricyclic guanines: A case study of dipeptidyl peptidase 4 (DPP4) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5T8W
 
 | |
5T4E
 
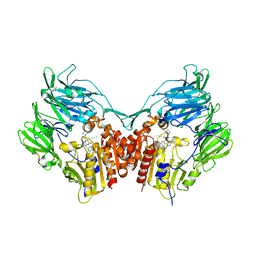 | | Human DPP4 in complex with ligand 19a | | Descriptor: | 2-[(3R)-3-aminopiperidin-1-yl]-3-(but-2-yn-1-yl)-6-[(4-methylquinazolin-2-yl)methyl]-6,7,8,9-tetrahydropyrimido[2,1-b]purin-4(3H)-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Scaffold-hopping from xanthines to tricyclic guanines: A case study of dipeptidyl peptidase 4 (DPP4) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5SWL
 
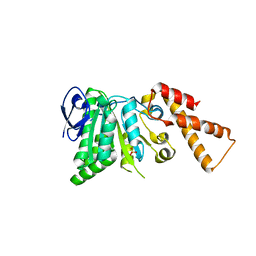 | | Crystal Structure of ATPase delta1-79 Spa47 E188A | | Descriptor: | Probable ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Burgess, J.L, Burgess, R.A, Dickenson, N.E. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Characterization of Spa47 Provides Mechanistic Insight into Type III Secretion System ATPase Activation and Shigella Virulence Regulation.
J. Biol. Chem., 291, 2016
|
|
1TR8
 
 | |
5SXV
 
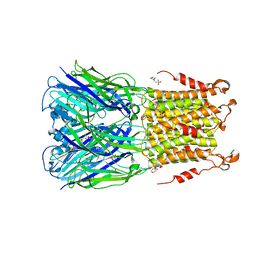 | | X-ray structure of 2-bromoethanol bound to a pentameric ligand gated ion channel (ELIC) in a resting state | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-BROMOETHANOL, Cys-loop ligand-gated ion channel | | Authors: | Chen, Q, Kinde, M, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2016-08-10 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Alcohol Inhibition of the Pentameric Ligand-Gated Ion Channel ELIC.
Structure, 25, 2017
|
|
1UBG
 
 | | MsREcA-dATP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, RecA | | Authors: | Datta, S, Krishna, R, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2003-04-04 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of Mycobacterium smegmatis RecA and Its Nucleotide Complexes
J.BACTERIOL., 185, 2003
|
|
5T25
 
 | | Kinetic, Spectral and Structural Characterization of the Slow Binding Inhibitor Acetopyruvate with Dihydrodipicolinate Synthase from Escherichia coli. | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, LYSINE, SODIUM ION | | Authors: | Chooback, L, Thomas, L.M, Karsten, W.E, Fleming, C.D, Seabourn, P. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Kinetic, Spectral and Structural Characterization of the Slow Binding Inhibitor Acetopyruvate with Dihydrodipicolinate Synthase from Escherichia coli.
To Be Published
|
|
1U3H
 
 | | Crystal structure of mouse TCR 172.10 complexed with MHC class II I-Au molecule at 2.4 A | | Descriptor: | H-2 class II histocompatibility antigen, A-U alpha chain, A-U beta chain, ... | | Authors: | Maynard, J, Petersson, K, Wilson, D.H, Adams, E.J, Blondelle, S.E, Boulanger, M.J, Wilson, D.B, Garcia, K.C. | | Deposit date: | 2004-07-21 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: insights into MHC bias and antigen specificity
Immunity, 22, 2005
|
|
1UFN
 
 | | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik) | | Descriptor: | putative nuclear protein homolog 5830484A20Rik | | Authors: | Tochio, N, Kobayashi, N, Koshiba, S, Kigawa, T, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik)
To be Published
|
|
1TNN
 
 | |
5SYP
 
 | | Crystal Structure of ATPase delta1-79 Spa47 K165A | | Descriptor: | Probable ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Burgess, J.L, Burgess, R.A, Dickenson, N.E. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Biochemical Characterization of Spa47 Provides Mechanistic Insight into Type III Secretion System ATPase Activation and Shigella Virulence Regulation.
J. Biol. Chem., 291, 2016
|
|
5T4F
 
 | | Human DPP4 in complex with ligand 34p | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-({2-[(3R)-3-aminopiperidin-1-yl]-3-(but-2-yn-1-yl)-4-oxo-3,4-dihydro-5H-imidazo[2,1-b]purin-5-yl}methyl)benzonitrile, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Scaffold-hopping from xanthines to tricyclic guanines: A case study of dipeptidyl peptidase 4 (DPP4) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5T73
 
 | |
1TTD
 
 | | SOLUTION-STATE STRUCTURE OF A DNA DODECAMER DUPLEX CONTAINING A CIS-SYN THYMINE CYCLOBUTANE DIMER | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*AP*TP*TP*CP*GP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*CP*GP*AP*AP*(TTD)P*AP*AP*G)-3') | | Authors: | Mcateer, K, Jing, Y, Kao, J, Taylor, J.-S, Kennedy, M.A. | | Deposit date: | 1999-01-20 | | Release date: | 1999-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution-state structure of a DNA dodecamer duplex containing a Cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA.
J.Mol.Biol., 282, 1998
|
|
1TTG
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE TENTH TYPE III MODULE OF FIBRONECTIN: AN INSIGHT INTO RGD-MEDIATED INTERACTIONS | | Descriptor: | FIBRONECTIN | | Authors: | Main, A.L, Harvey, T.S, Baron, M, Campbell, I.D. | | Deposit date: | 1993-07-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions.
Cell(Cambridge,Mass.), 71, 1992
|
|
5T7M
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-TRP | | Descriptor: | ACETATE ION, Chemotaxis protein, SODIUM ION, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
1U22
 
 | | A. thaliana cobalamine independent methionine synthase | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, ... | | Authors: | Ferrer, J.-L, Ravanel, S, Robert, M, Dumas, R. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of cobalamin-independent methionine synthase complexed with zinc, homocysteine, and methyltetrahydrofolate
J.Biol.Chem., 279, 2004
|
|
