7DRD
 
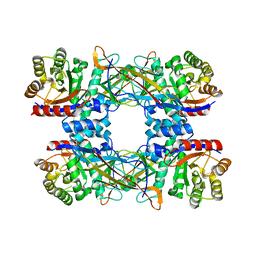 | | Cryo-EM structure of DgpB-C at 2.85 angstrom resolution | | Descriptor: | AP_endonuc_2 domain-containing protein, DgpB | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7EA8
 
 | |
6IJZ
 
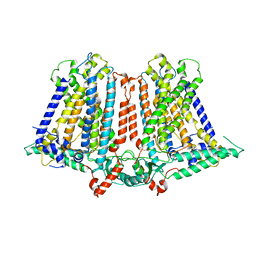 | | Structure of a plant cation channel | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Sun, L, Wang, J, Liu, X. | | Deposit date: | 2018-10-12 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2.
Nat Commun, 9, 2018
|
|
6AP5
 
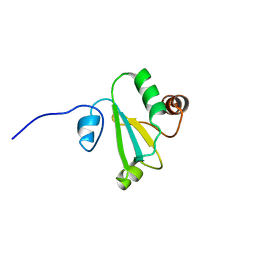 | | H, 13C, and 15N Chemical Shift Assignments and structure of Thioredoxin from Mycobacterium thermoresistibile ATCC 19527 and NCTC 10409 | | Descriptor: | Thioredoxin | | Authors: | Tang, C.T, Yang, F.Y, Varani, G.V, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2017-08-17 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | H, 13C, and 15N Chemical Shift Assignments and structure of Thioredoxin from Mycobacterium thermoresistibile ATCC 19527 and NCTC 10409
To Be Published
|
|
6B2X
 
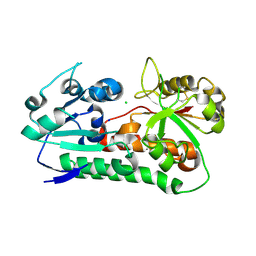 | | Apo YiuA Crystal Form 1 | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute-binding periplasmic protein of iron/siderophore ABC transporter | | Authors: | Radka, C.D, DeLucas, L.J, Aller, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The crystal structure of the Yersinia pestis iron chaperone YiuA reveals a basic triad binding motif for the chelated metal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6IAT
 
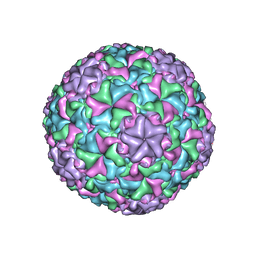 | | Icosahedrally averaged capsid of bacteriophage P68 | | Descriptor: | Arstotzka protein, Major head protein | | Authors: | Hrebik, D, Skubnik, K, Fuzik, T, Plevka, P. | | Deposit date: | 2018-11-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
6I2N
 
 | | Helical RNA-bound Hantaan virus nucleocapsid | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*U)-3') | | Authors: | Arragain, B, Reguera, J, Desfosses, A, Gutsche, I, Schoehn, G, Malet, H. | | Deposit date: | 2018-11-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High resolution cryo-EM structure of the helical RNA-bound Hantaan virus nucleocapsid reveals its assembly mechanisms.
Elife, 8, 2019
|
|
4Y4J
 
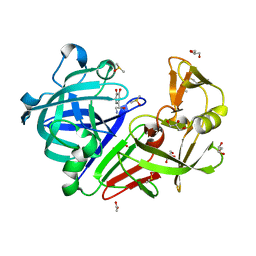 | | Endothiapepsin in complex with fragment B97 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6V8O
 
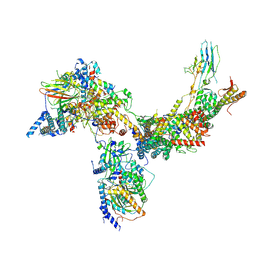 | | RSC core | | Descriptor: | Chromatin structure-remodeling complex protein RSC14, Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, ... | | Authors: | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
4Y4D
 
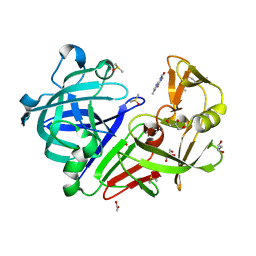 | | Endothiapepsin in complex with fragment B51 | | Descriptor: | ACETATE ION, CAFFEINE, DIMETHYL SULFOXIDE, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2016-05-11 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
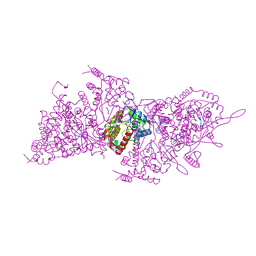
6H05
 
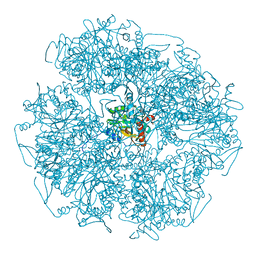 | | Cryo-electron microscopic structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate (2-oxoglutarate) dehydrogenase complex [residues 218-453] | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | | Authors: | Nagy, B, Zambo, Z, Hubert, A, Polak, M, Nemeria, N.S, Novacek, J, Jordan, F, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-07-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate dehydrogenase complex (hKGDHc) revealed by cryo-EM and cross-linking mass spectrometry: Implications for the overall hKGDHc structure.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
7E8D
 
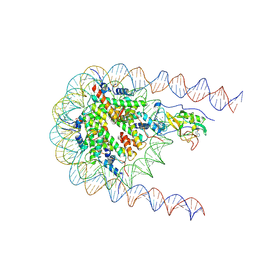 | | NSD2 E1099K mutant bound to nucleosome | | Descriptor: | DNA (185-MER), Histone H2A type 1, Histone H2B type 1-J, ... | | Authors: | Sengoku, T, Sato, K, Nishizawa, T, Nureki, O, Ogata, K. | | Deposit date: | 2021-03-01 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of the regulation of the normal and oncogenic methylation of nucleosomal histone H3 Lys36 by NSD2.
Nat Commun, 12, 2021
|
|
7EH7
 
 | | Cryo-EM structure of the octameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Adachi, N, Hung, N.K, Yamada, K, Kawasaki, M, Akutsu, M, Moriya, T, Senda, T, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
7EH8
 
 | | Cryo-EM structure of the hexameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Adachi, N, Hung, N.K, Yamada, K, Kawasaki, M, Akutsu, M, Moriya, T, Senda, T, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
6I7T
 
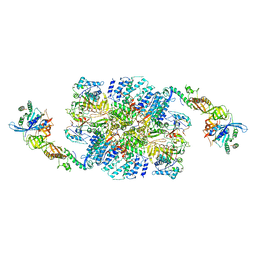 | | eIF2B:eIF2 complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Adomavicius, T, Guaita, M, Roseman, A.M, Pavitt, G.D. | | Deposit date: | 2018-11-17 | | Release date: | 2019-05-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.61 Å) | | Cite: | The structural basis of translational control by eIF2 phosphorylation.
Nat Commun, 10, 2019
|
|
6B2Y
 
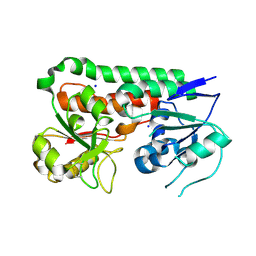 | | Apo YiuA Crystal Form 2 | | Descriptor: | SODIUM ION, Solute-binding periplasmic protein of iron/siderophore ABC transporter | | Authors: | Radka, C.D, DeLucas, L.J, Aller, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The crystal structure of the Yersinia pestis iron chaperone YiuA reveals a basic triad binding motif for the chelated metal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8UO1
 
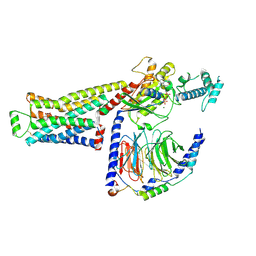 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class Q) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
7EJE
 
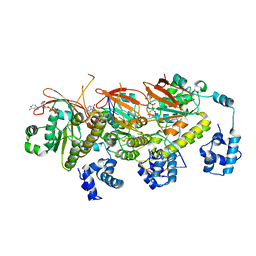 | | human RAD51 post-synaptic complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Zhao, L.Y, Xu, J.F, Wang, H.W. | | Deposit date: | 2021-04-02 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
7EJC
 
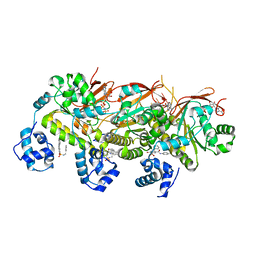 | | human RAD51 presynaptic complex | | Descriptor: | 4-bromanyl-N-(4-bromophenyl)-3-[(phenylmethyl)sulfamoyl]benzamide, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Zhao, L.Y, Xu, J.F, Wang, H.W. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
6BF2
 
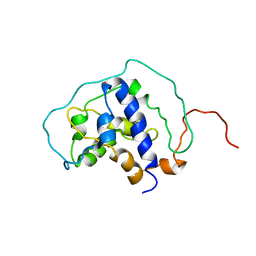 | |
6TZ9
 
 | |
4WNZ
 
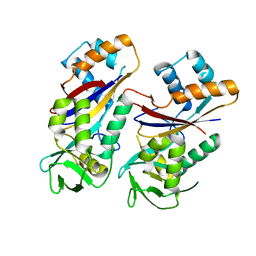 | |
7E80
 
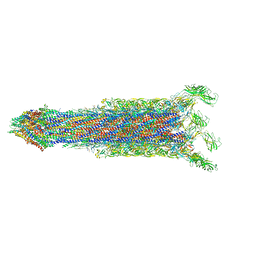 | | Cryo-EM structure of the flagellar rod with hook and export apparatus from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7E82
 
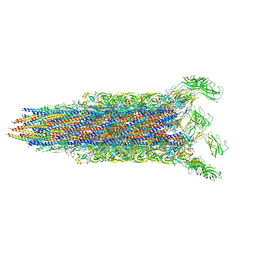 | | Cryo-EM structure of the flagellar rod with partial hook from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7DCR
 
 | | cryo-EM structure of the DEAH-box helicase Prp2 in complex with its coactivator Spp2 | | Descriptor: | PRP2 isoform 1, Pre-mRNA-splicing factor SPP2 | | Authors: | Bai, R, Wan, R, Yan, C, Jia, Q, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-26 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
