5FEL
 
 | |
5FEZ
 
 | | HydE from T. maritima in complex with (2R,4R)-MeSeTDA, 5'-deoxyadenosine and methionine | | Descriptor: | (2~{R},4~{R})-2-methyl-1,3-selenazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
5X3K
 
 | | Kfla1895 D451A mutant in complex with isomaltose | | Descriptor: | GLYCEROL, Glycoside hydrolase family 31, SULFATE ION, ... | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glycoside hydrolase mutant in complex with product
To Be Published
|
|
2VO5
 
 | | Structural and biochemical evidence for a boat-like transition state in beta-mannosidases | | Descriptor: | (1R,4R,5R,7R,8R)-2-Benzyl-5-hydroxymethyl-2-aza-bicyclo[2.2.2]octane-4,7,8-triol, 1,2-ETHANEDIOL, BETA-MANNOSIDASE, ... | | Authors: | Tailford, L.E, Offen, W.A, Smith, N.L, Dumon, C, Moreland, C, Gratien, J, Heck, M.P, Stick, R.V, Bleriot, Y, Vasella, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Evidence for a Boat-Like Transition State in Beta-Mannosidases.
Nat.Chem.Biol., 4, 2008
|
|
7N8O
 
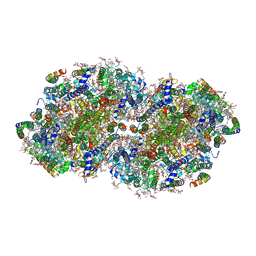 | | High-resolution structure of photosystem II from the mesophilic cyanobacterium, Synechocystis sp. PCC 6803 | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Gisriel, C.J, Brudvig, G.W. | | Deposit date: | 2021-06-15 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | High-resolution cryo-electron microscopy structure of photosystem II from the mesophilic cyanobacterium, Synechocystis sp. PCC 6803.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8HBA
 
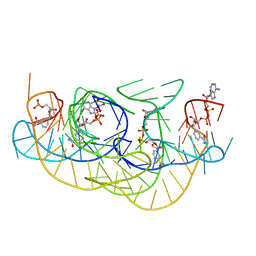 | |
5XVQ
 
 | | Crystal structure of monkey Nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl Nicotinamide (MNA) | | Descriptor: | 3-carbamoyl-1-methylpyridin-1-ium, GLYCEROL, Nicotinamide N-methyltransferase (NNMT), ... | | Authors: | Birudukota, S, Swaminathan, S, Thakur, M.K, Parveen, R, Kandan, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of monkey and mouse nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl nicotinamide
Biochem. Biophys. Res. Commun., 491, 2017
|
|
7NGE
 
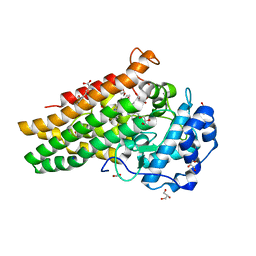 | | Crystal structure of L-Trp/Indoleamine 2,3-dioxygenagse 1 (hIDO1) complex with the JK-loop refined in the closed conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Mirgaux, M, Wouters, J. | | Deposit date: | 2021-02-09 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Temporary Intermediates of L-Trp Along the Reaction Pathway of Human Indoleamine 2,3-Dioxygenase 1 and Identification of an Exo Site.
Int J Tryptophan Res, 14, 2021
|
|
5XVV
 
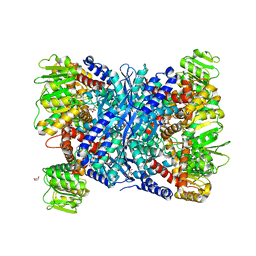 | | Crystal Structure of Forward Inhibited Aspergillus niger Glutamate Dehydrogenase With Both Apo- and Alpha Ketoglutarate Bound Subunits | | Descriptor: | 2-OXOGLUTARIC ACID, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Prakash, P, Punekar, N.S, Bhaumik, P. | | Deposit date: | 2017-06-28 | | Release date: | 2018-03-21 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the catalytic mechanism and alpha-ketoglutarate cooperativity of glutamate dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
8HB8
 
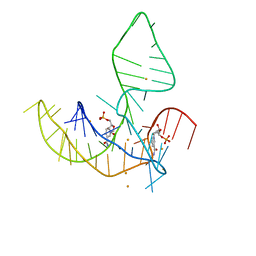 | |
8HB3
 
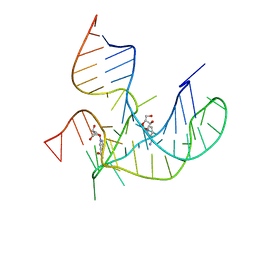 | | Crystal structure of NAD-II riboswitch (two strands) with NR | | Descriptor: | Nicotinamide riboside, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3') | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
6SYC
 
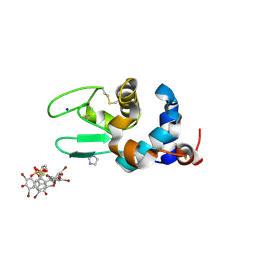 | | Crystal structure of the lysozyme in presence of bromophenol blue at pH 6.5 | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Lysozyme, ... | | Authors: | Camara-Artigas, A, Plaza-Garrido, M, Salinas-Garcia, M.C. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Lysozyme crystals dyed with bromophenol blue: where has the dye gone?
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6T0L
 
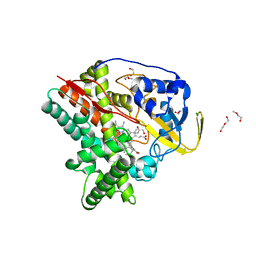 | | Crystal structure of CYP124 in complex with inhibitor compound 5' | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor compound 5', DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
3P5H
 
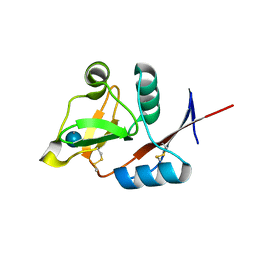 | | Structure of the carbohydrate-recognition domain of human Langerin with Laminaritriose | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, beta-D-glucopyranose, ... | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6051 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
3NML
 
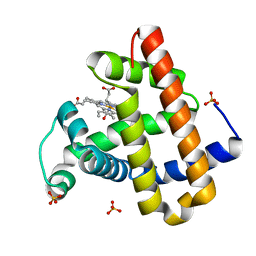 | | Sperm whale myoglobin mutant H64W carbonmonoxy-form | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Birukou, I, Soman, J, Olson, J.S. | | Deposit date: | 2010-06-22 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Blocking the gate to ligand entry in human hemoglobin.
J.Biol.Chem., 286, 2011
|
|
2VUE
 
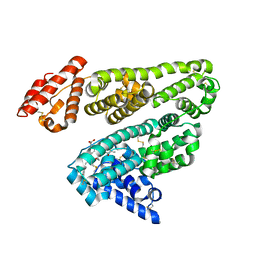 | | Human serum albumin complexed with 4Z,15E-bilirubin-IX-alpha | | Descriptor: | BILIVERDINE IX ALPHA, SERUM ALBUMIN | | Authors: | Zunszain, P.A, Ghuman, J, Curry, S. | | Deposit date: | 2008-05-24 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystallographic Analysis of Human Serum Albumin Complexed with 4Z,15E-Bilirubin-Ixalpha.
J.Mol.Biol., 381, 2008
|
|
8HJA
 
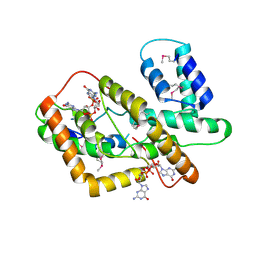 | | The crystal structure of syn_CdgR-(c-di-GMP) from Synechocystis sp. PCC 6803 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), c-di-GMP receptor | | Authors: | Zeng, X, Peng, Y.J. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A c-di-GMP binding effector controls cell size in a cyanobacterium.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1UMA
 
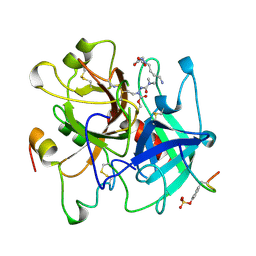 | | ALPHA-THROMBIN (HIRUGEN) COMPLEXED WITH NA-(N,N-DIMETHYLCARBAMOYL)-ALPHA-AZALYSINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUDIN I, ... | | Authors: | Nardini, M, Pesce, A, Rizzi, M, Casale, E, Ferraccioli, R, Balliano, G, Milla, P, Ascenzi, P, Bolognesi, M. | | Deposit date: | 1996-03-26 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the active site titrant N alpha-(N,N-dimethylcarbamoyl)-alpha-azalysine p-nitrophenyl ester: a comparative kinetic and X-ray crystallographic study.
J.Mol.Biol., 258, 1996
|
|
6VCQ
 
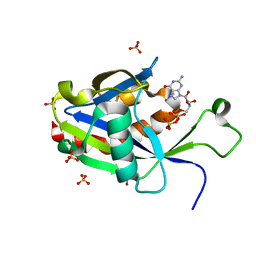 | | Crystal structure of E.coli RppH in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA pyrophosphohydrolase, SULFATE ION | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
5XDE
 
 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and dibenzothiophene | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
1KWJ
 
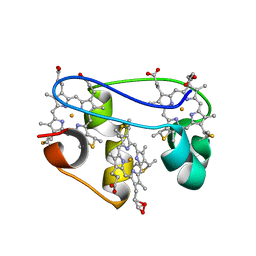 | | solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans, minimized average structure | | Descriptor: | HEME C, cytochrome c7 | | Authors: | Assfalg, M, Bertini, I, Turano, P, Bruschi, M, Durand, M.C, Giudici-Orticoni, M.T, Dolla, A. | | Deposit date: | 2002-01-29 | | Release date: | 2002-02-06 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A quick solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans and mechanistic implications.
J.Biomol.NMR, 22, 2002
|
|
7N14
 
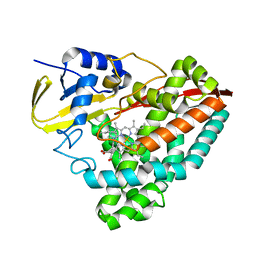 | | Crystal structure of 4-(1H-1,2,4-triazol-1-yl)benzoic acid-bound CYP199A4 | | Descriptor: | 4-(1H-1,2,4-triazol-1-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2021-05-26 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | To Be, or Not to Be, an Inhibitor: A Comparison of Azole Interactions with and Oxidation by a Cytochrome P450 Enzyme.
Inorg.Chem., 61, 2022
|
|
2VXX
 
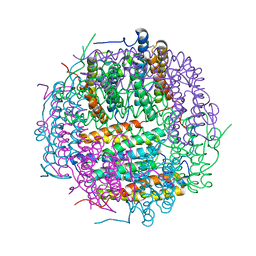 | | X-ray structure of DpsA from Thermosynechococcus elongatus | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, STARVATION INDUCED DNA BINDING PROTEIN, ... | | Authors: | Franceschini, S, Ilari, A. | | Deposit date: | 2008-07-14 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermosynechococcus Elongatus Dpsa Binds Zn(II) at a Unique Three Histidine-Containing Ferroxidase Center and Utilizes O2 as Iron Oxidant with Very High Efficiency, Unlike the Typical Dps Proteins.
FEBS J., 277, 2010
|
|
1KX8
 
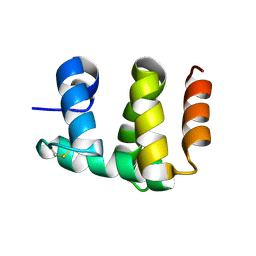 | | Antennal Chemosensory Protein A6 from Mamestra brassicae, tetragonal form | | Descriptor: | CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Ligand Binding Study of a Chemosensory Protein
J.Biol.Chem., 277, 2002
|
|
1KDP
 
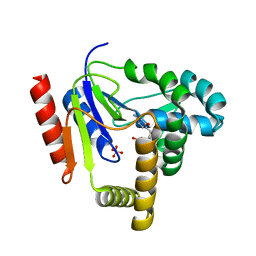 | | CYTIDINE MONOPHOSPHATE KINASE FROM E. COLI IN COMPLEX WITH 2'-DEOXY-CYTIDINE MONOPHOSPHATE | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CYTIDYLATE KINASE, SULFATE ION | | Authors: | Bertrand, T, Briozzo, P, Assairi, L, Ofiteru, A, Bucurenci, N, Munier-Lehmann, H, Golinelli-Pimpaneau, B, Barzu, O, Gilles, A.M. | | Deposit date: | 2001-11-13 | | Release date: | 2002-01-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sugar specificity of bacterial CMP kinases as revealed by crystal structures and mutagenesis of Escherichia coli enzyme.
J.Mol.Biol., 315, 2002
|
|
