4TUF
 
 | |
6OMF
 
 | | CryoEM structure of SigmaS-transcription initiation complex with activator Crl | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Jaramillo Cartagena, A, Darst, S.A, Campbell, E.A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for transcription activation by Crl through tethering of sigmaSand RNA polymerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6M7J
 
 | | Mycobacterium tuberculosis RNAP with RbpA/us fork and Corallopyronin | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-20 | | Release date: | 2018-11-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
4TR6
 
 | | Crystal structure of DNA polymerase sliding clamp from Bacillus subtilis | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION | | Authors: | Burnouf, D, Olieric, V, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-14 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
3BEP
 
 | | Structure of a sliding clamp on DNA | | Descriptor: | 1-(3-hydroxypropyl)-2-{(1E,3E,5E)-5-[1-(3-hydroxypropyl)-3,3-dimethyl-1,3-dihydro-2H-indol-2-ylidene]penta-1,3-dien-1-y l}-3,3-dimethyl-3H-indolium, DNA (5'-D(*DTP*DTP*DTP*DTP*DAP*DTP*DAP*DCP*DGP*DAP*DTP*DGP*DGP*DG)-3'), DNA (5'-D(P*DCP*DCP*DCP*DAP*DTP*DCP*DGP*DTP*DAP*DT)-3'), ... | | Authors: | Georgescu, R.E, Kim, S.S, Yurieva, O, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2007-11-19 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of a sliding clamp on DNA
Cell(Cambridge,Mass.), 132, 2008
|
|
3FIE
 
 | |
4TSZ
 
 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa with ligand | | Descriptor: | ACE-GLN-ALC-ASP-LEU-ZCL peptide, DNA polymerase III subunit beta | | Authors: | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-19 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
2QCS
 
 | | A complex structure between the Catalytic and Regulatory subunit of Protein Kinase A that represents the inhibited state | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kim, C, Cheng, C.Y, Saldanha, A.S, Taylor, S.S. | | Deposit date: | 2007-06-19 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation.
Cell(Cambridge,Mass.), 130, 2007
|
|
1JQL
 
 | | Mechanism of Processivity Clamp Opening by the Delta Subunit Wrench of the Clamp Loader Complex of E. coli DNA Polymerase III: Structure of beta-delta (1-140) | | Descriptor: | DNA Polymerase III, BETA CHAIN, DELTA SUBUNIT | | Authors: | Jeruzalmi, D, Yurieva, O, Zhao, Y, Young, M, Stewart, J, Hingorani, M, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
1UC3
 
 | |
8XVN
 
 | |
8XVO
 
 | |
6N57
 
 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound with TraR in conformation I | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-21 | | Release date: | 2020-02-26 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
6N58
 
 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound with TraR in conformation II | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-21 | | Release date: | 2020-02-26 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
5CDE
 
 | | R372A mutant of Xaa-Pro dipeptidase from Xanthomonas campestris | | Descriptor: | Proline dipeptidase, SULFATE ION, ZINC ION | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-03 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | R372A mutant of Xaa-Pro dipeptidase from Xanthomonas campestris at 1.85 Angstrom resolution
To Be Published
|
|
6PTV
 
 | |
6AMS
 
 | |
5CIK
 
 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris in citrate condition | | Descriptor: | CITRIC ACID, GLYCEROL, Proline dipeptidase | | Authors: | Kumar, A, Are, V, Ghosh, B, Agrawal, U, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-07-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris in citrate condition
To Be Published
|
|
6PTH
 
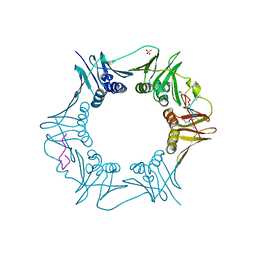 | |
4AKD
 
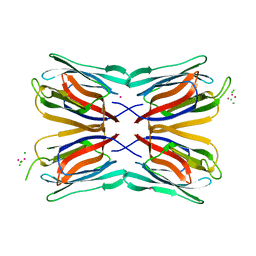 | | High resolution structure of Mannose Binding lectin from Champedak (CMB) | | Descriptor: | CADMIUM ION, CHLORIDE ION, MANNOSE-SPECIFIC LECTIN KM+ | | Authors: | Gabrielsen, M, Abdul-Rahman, P.S, Othman, S, Hashim, O.H, Cogdell, R.J. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and Binding Specificity of Galactose- and Mannose-Binding Lectins from Champedak: Differences from Jackfruit Lectins
Acta Crystallogr.,Sect.F, 70, 2014
|
|
6S4G
 
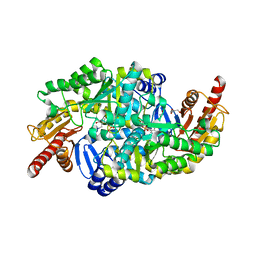 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruggieri, F, Campillo Brocal, J.C, Humble, M.S, Walse, B, Logan, D.T, Berglund, P. | | Deposit date: | 2019-06-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Insight into the dimer dissociation process of the Chromobacterium violaceum (S)-selective amine transaminase.
Sci Rep, 9, 2019
|
|
3UIB
 
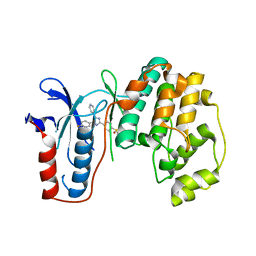 | | Map kinase LMAMPK10 from leishmania major in complex with SB203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, mitogen-activated protein kinase | | Authors: | Horjales, S, Schmidt-Arras, D, Leclercq, O, Spath, G, Buschiazzo, A. | | Deposit date: | 2011-11-04 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of the MAP Kinase LmaMPK10 from Leishmania Major Reveals Parasite-Specific Features and Regulatory Mechanisms.
Structure, 20, 2012
|
|
3TAZ
 
 | | Crystal structure of NurA bound to dAMP and manganese | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA double-strand break repair protein nurA, GLYCEROL, ... | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
3DMB
 
 | |
1X3N
 
 | | Crystal structure of AMPPNP bound Propionate kinase (TdcD) from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Propionate kinase | | Authors: | Simanshu, D.K, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of ADP and AMPPNP-bound Propionate Kinase (TdcD) from Salmonella typhimurium: Comparison with Members of Acetate and Sugar Kinase/Heat Shock Cognate 70/Actin Superfamily.
J.Mol.Biol., 352, 2005
|
|
