6Q0Y
 
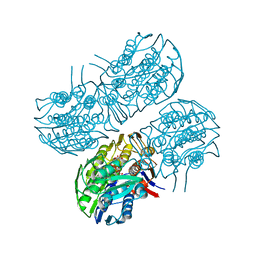 | | Crystal structure of MurA from Clostridium difficile, mutant C116S, in the presence of Uridine-Diphosphate-N-Acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutant C116S, in the presence of Uridine-Diphosphate-N-Acetylglucosamine
To Be Published
|
|
6Y87
 
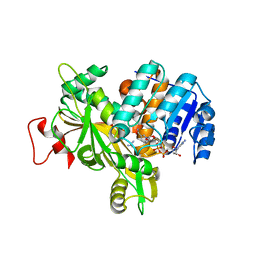 | |
8EXG
 
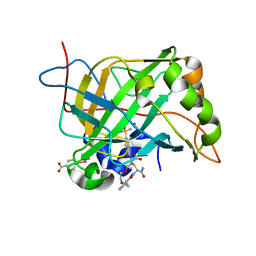 | | Human Carbonic Anhydrase II bound N-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethyl)-4-sulfamoylbenzamide | | Descriptor: | (4S)-5-amino-4-{1,3-dioxo-4-[(2-{2-[2-(4-sulfamoylbenzamido)ethoxy]ethoxy}ethyl)amino]-1,3-dihydro-2H-isoindol-2-yl}-5-oxopentanoic acid, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
8KEE
 
 | | Cyanophage A-1(L) sheath-tube | | Descriptor: | sheath, tube | | Authors: | Yu, R.C, Li, Q, Zhou, C.Z. | | Deposit date: | 2023-08-11 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the intact tail machine of Anabaena myophage A-1(L).
Nat Commun, 15, 2024
|
|
8R2E
 
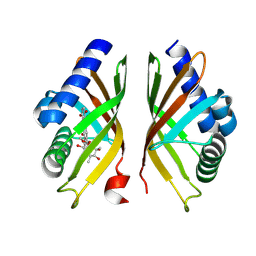 | |
8F2P
 
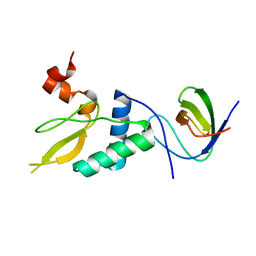 | |
6YH9
 
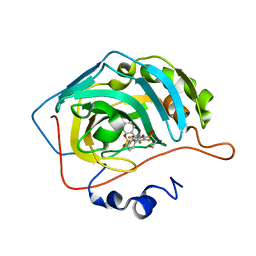 | | Crystal structure of chimeric carbonic anhydrase XII with 2,3,6-trifluoro-5-{[(1R,2S)-2-hydroxy-1,2-diphenylethyl]amino}-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 2,3,6-trifluoro-5-{[(1R,2S)-2-hydroxy-1,2-diphenylethyl]amino}-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
6YH7
 
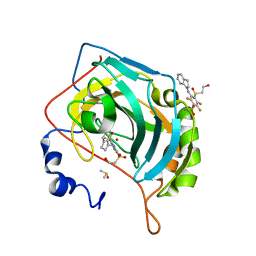 | | Crystal structure of chimeric carbonic anhydrase XII with 3-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 3-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
7OZB
 
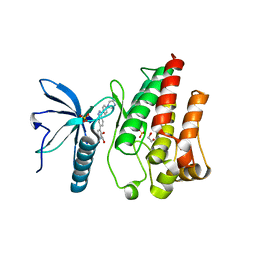 | | FGFR1 kinase domain (residues 458-765) with mutations C488A, C584S in complex with 38. | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-(4-piperazin-4-ium-1-ylphenyl)-1H-indazol-6-yl]phenol, Fibroblast growth factor receptor 1, ... | | Authors: | Trinh, C.H, Turner, L.D, Fishwick, C.W.G. | | Deposit date: | 2021-06-27 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | From Fragment to Lead: De Novo Design and Development toward a Selective FGFR2 Inhibitor.
J.Med.Chem., 65, 2022
|
|
8KE9
 
 | |
4X59
 
 | | Anthranilate phosphoribosyltransferase variant P180A from Mycobacterium tuberculosis in complex with PRPP and Mg | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, GLYCEROL, ... | | Authors: | Cookson, T.V.M, Evans, G.L, Parker, E.J, Lott, J.S. | | Deposit date: | 2014-12-04 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Variants Reveal the Conformational Changes That Facilitate Delivery of the Substrate to the Active Site.
Biochemistry, 54, 2015
|
|
6W4R
 
 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ633p | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-{[2-(2-hydroxyphenyl)imidazo[1,2-a]pyrazin-3-yl]amino}benzene-1,2-dicarboxylic acid, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Tdp1 catalytic domain
To Be Published
|
|
7ASL
 
 | |
4X6O
 
 | |
8TCT
 
 | | Structure of 3K-GlcH bound Bacteroides thetaiotaomicron 3-Keto-beta-glucopyranoside-1,2-Lyase BT1 | | Descriptor: | 1,5-anhydro-D-ribo-hex-3-ulose, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
6Q8B
 
 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with 5'-O-(N-(L-Leucyl)-Sulfamoyl)N3-methyluridine | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative analysis of pyrimidine substituted aminoacyl-sulfamoyl nucleosides as potential inhibitors targeting class I aminoacyl-tRNA synthetases.
Eur.J.Med.Chem., 173, 2019
|
|
6W7J
 
 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ635p | | Descriptor: | 1,2-ETHANEDIOL, 4-{[2-(2-hydroxyphenyl)imidazo[1,2-a]pyridin-3-yl]amino}benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Tdp1 catalytic domain
To Be Published
|
|
4WL4
 
 | |
4WYG
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis complexed with a fragment hit | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[(4-chloro-1H-pyrazol-1-yl)methyl]phenyl}methanamine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Dai, R, Finzel, B.C. | | Deposit date: | 2014-11-17 | | Release date: | 2015-07-08 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fragment-Based Exploration of Binding Site Flexibility in Mycobacterium tuberculosis BioA.
J.Med.Chem., 58, 2015
|
|
6O1S
 
 | | Structure of human plasma kallikrein protease domain with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-[(6-amino-2,4-dimethylpyridin-3-yl)methyl]-1-({4-[(1H-pyrazol-1-yl)methyl]phenyl}methyl)-1H-pyrazole-4-carboxamide, PHOSPHATE ION, ... | | Authors: | Partridge, J.R, Choy, R.M. | | Deposit date: | 2019-02-21 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of full-length plasma kallikrein bound to highly specific inhibitors describe a new mode of targeted inhibition.
J.Struct.Biol., 206, 2019
|
|
6WAK
 
 | | A crystal structure of EGFR(T790M/V948R) in complex with LN3754 | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-(3-{5-[2-(acetylamino)pyridin-4-yl]-2-(methylsulfanyl)-1H-imidazol-4-yl}phenyl)-2-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]benzamide, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
8RSI
 
 | | Crystal structure of Methanobrevibacter oralis macrodomain | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTAMIC ACID, ... | | Authors: | Ariza, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains.
J.Biol.Chem., 300, 2024
|
|
6QEF
 
 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX WITH AN INHIBITOR (S)-3-Hydroxy-2-oxo-1-phenyl-pyrrolidine-3-carboxylic acid 3-chloro-5-fluoro-benzylamide | | Descriptor: | (3~{S})-~{N}-[(3-chloranyl-5-fluoranyl-phenyl)methyl]-3-oxidanyl-2-oxidanylidene-1-phenyl-pyrrolidine-3-carboxamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
8AI1
 
 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-homocysteine bound. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Polsinelli, I, Chavas, L.M.G, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
6NXC
 
 | |
